Abstract
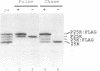
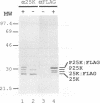
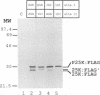
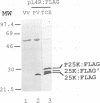
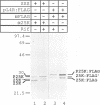
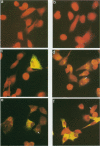
The three major vaccinia virus (VV) virion proteins (4a, 4b, and 25K) are proteolytically matured from larger precursors (P4a, P4b, and P25K) during virus assembly. Within the precursors, Ala-Gly-X motifs have been noted at the putative processing sites, with cleavage apparently taking place between the Gly and X residues. To identify the sequence and/or structural parameters which are required to define an efficient cleavage site, a trans-processing assay system has been developed by tagging the carboxy terminus of the P25K polypeptide (precursor of 25K) with an octapeptide FLAG epitope, which can be specifically recognized by a monoclonal antibody. By using transient expression assays with cells coinfected with VV, the proteolytic processing of the chimeric gene product (P25K:FLAG) was monitored by immunoblotting procedures. The relationship between the P25K:FLAG precursor and the 25K:FLAG cleavage product was established by pulse-chase experiments. The in vivo cleavage of P25K:FLAG was inhibited by the drug rifampin, implying that the reaction was utilizing the same pathway as authentic VV core proteins. Moreover, the 25K:FLAG protein was found in association with mature virions in accord with the notion that cleavage occurs concomitantly with virion assembly. Site-directed mutagenesis of the Ala-Gly-Ala motif at residues 31 to 33 of the P25K:FLAG precursor to Ile-Asp-Ile blocked production of the 25K:FLAG product. The efficiency of 25K:FLAG production (33.71%) is, however, approximately only half of the production of 25K (63.98%) within VV-infected cells transfected with pL4R:FLAG. One explanation for the lower efficiency of 25K:FLAG production was suggested by the observation in the immunofluorescent-staining experiment that 25K:FLAG-related proteins were not specifically localized to the virus assembly factories (virosomes) within VV-infected cells, although virosome localization was prominent for P25K-related polypeptides. Since VV core protein proteolytic processing is believed to take place during virion maturation, only the P25K:FLAG which was assembled into immature virions could undergo proteolytic maturation. Furthermore during these experiments, a potential cleavage intermediate (25K') of P25K was identified. Amino acid residues 17 to 19 (Ala-Gly-Ser) of the P25K precursor were implicated as the intermediate cleavage site, since no 25K':FLAG product was produced from a mutant precursor in which the sequence was altered to Ile-Asp-Ile. Taken together, these results provide biochemical and genetic evidence to support the hypothesis that the Ala-Gly-X cleavage motif plays a critical role in VV virion protein proteolytic maturation.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akita M., Sasaki S., Matsuyama S., Mizushima S. SecA interacts with secretory proteins by recognizing the positive charge at the amino terminus of the signal peptide in Escherichia coli. J Biol Chem. 1990 May 15;265(14):8164–8169. [PubMed] [Google Scholar]
- Ashorn P., McQuade T. J., Thaisrivongs S., Tomasselli A. G., Tarpley W. G., Moss B. An inhibitor of the protease blocks maturation of human and simian immunodeficiency viruses and spread of infection. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7472–7476. doi: 10.1073/pnas.87.19.7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns M. M., Boursnell M. E., Tomley F. M., Campbell J. Analysis of the fowlpoxvirus gene encoding the 4b core polypeptide and demonstration that it possesses efficient promoter sequences. Virology. 1989 May;170(1):288–291. doi: 10.1016/0042-6822(89)90380-2. [DOI] [PubMed] [Google Scholar]
- Binns M. M., Tomley F. M., Campbell J., Boursnell M. E. Comparison of a conserved region in fowlpox virus and vaccinia virus genomes and the translocation of the fowlpox virus thymidine kinase gene. J Gen Virol. 1988 Jun;69(Pt 6):1275–1283. doi: 10.1099/0022-1317-69-6-1275. [DOI] [PubMed] [Google Scholar]
- Chou M. M., Kendall D. A. Polymeric sequences reveal a functional interrelationship between hydrophobicity and length of signal peptides. J Biol Chem. 1990 Feb 15;265(5):2873–2880. [PubMed] [Google Scholar]
- Condit R. C., Motyczka A., Spizz G. Isolation, characterization, and physical mapping of temperature-sensitive mutants of vaccinia virus. Virology. 1983 Jul 30;128(2):429–443. doi: 10.1016/0042-6822(83)90268-4. [DOI] [PubMed] [Google Scholar]
- Dales S., Milovanovitch V., Pogo B. G., Weintraub S. B., Huima T., Wilton S., McFadden G. Biogenesis of vaccinia: isolation of conditional lethal mutants and electron microscopic characterization of their phenotypically expressed defects. Virology. 1978 Feb;84(2):403–428. doi: 10.1016/0042-6822(78)90258-1. [DOI] [PubMed] [Google Scholar]
- Dougherty W. G., Cary S. M., Parks T. D. Molecular genetic analysis of a plant virus polyprotein cleavage site: a model. Virology. 1989 Aug;171(2):356–364. doi: 10.1016/0042-6822(89)90603-x. [DOI] [PubMed] [Google Scholar]
- Dyster L. M., Niles E. G. Genetic and biochemical characterization of vaccinia virus genes D2L and D3R which encode virion structural proteins. Virology. 1991 Jun;182(2):455–467. doi: 10.1016/0042-6822(91)90586-z. [DOI] [PubMed] [Google Scholar]
- Fathi Z., Condit R. C. Genetic and molecular biological characterization of a vaccinia virus temperature-sensitive complementation group affecting a virion component. Virology. 1991 Mar;181(1):258–272. doi: 10.1016/0042-6822(91)90491-s. [DOI] [PubMed] [Google Scholar]
- Fathi Z., Condit R. C. Phenotypic characterization of a vaccinia virus temperature-sensitive complementation group affecting a virion component. Virology. 1991 Mar;181(1):273–276. doi: 10.1016/0042-6822(91)90492-t. [DOI] [PubMed] [Google Scholar]
- Fuerst T. R., Niles E. G., Studier F. W., Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T. M., Rueckert R. R. Assembly-dependent maturation cleavage in provirions of a small icosahedral insect ribovirus. J Virol. 1988 Sep;62(9):3399–3406. doi: 10.1128/jvi.62.9.3399-3406.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel S. J., Johnson G. P., Perkus M. E., Davis S. W., Winslow J. P., Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990 Nov;179(1):247-66, 517-63. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- Harmar A. J., Keen P. M. Methods for the identification of neuropeptide processing products: somatostatin and the tachykinins. Methods Enzymol. 1986;124:335–348. doi: 10.1016/0076-6879(86)24025-2. [DOI] [PubMed] [Google Scholar]
- Hellen C. U., Wimmer E. Maturation of poliovirus capsid proteins. Virology. 1992 Apr;187(2):391–397. doi: 10.1016/0042-6822(92)90440-z. [DOI] [PubMed] [Google Scholar]
- Hruby D. E., Guarino L. A., Kates J. R. Vaccinia virus replication. I. Requirement for the host-cell nucleus. J Virol. 1979 Feb;29(2):705–715. doi: 10.1128/jvi.29.2.705-715.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y., Oie M., Tsuruhara T. Location of DNA-binding proteins and disulfide-linked proteins in vaccinia virus structural elements. J Virol. 1984 Jun;50(3):929–938. doi: 10.1128/jvi.50.3.929-938.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung L. J., Scheller R. H. Peptide processing and targeting in the neuronal secretory pathway. Science. 1991 Mar 15;251(4999):1330–1335. doi: 10.1126/science.2003219. [DOI] [PubMed] [Google Scholar]
- Katz E., Moss B. Formation of a vaccinia virus structural polypeptide from a higher molecular weight precursor: inhibition by rifampicin. Proc Natl Acad Sci U S A. 1970 Jul;66(3):677–684. doi: 10.1073/pnas.66.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989 Feb;108(2):229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kräusslich H. G., Wimmer E. Viral proteinases. Annu Rev Biochem. 1988;57:701–754. doi: 10.1146/annurev.bi.57.070188.003413. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lake J. R., Silver M., Dales S. Biogenesis of vaccinia: complementation and recombination analysis of one group of conditional-lethal mutants defective in envelope self-assembly. Virology. 1979 Jul 15;96(1):9–20. doi: 10.1016/0042-6822(79)90167-3. [DOI] [PubMed] [Google Scholar]
- Lawson M. A., Semler B. L. Picornavirus protein processing--enzymes, substrates, and genetic regulation. Curr Top Microbiol Immunol. 1990;161:49–87. [PubMed] [Google Scholar]
- López-Otín C., Simón-Mateo C., Martínez L., Viñuela E. Gly-Gly-X, a novel consensus sequence for the proteolytic processing of viral and cellular proteins. J Biol Chem. 1989 Jun 5;264(16):9107–9110. [PubMed] [Google Scholar]
- Miner J. N., Hruby D. E. Rifampicin prevents virosome localization of L65, an essential vaccinia virus polypeptide. Virology. 1989 May;170(1):227–237. doi: 10.1016/0042-6822(89)90370-x. [DOI] [PubMed] [Google Scholar]
- Moss B., Rosenblum E. N., Katz E., Grimley P. M. Rifampicin: a specific inhibitor of vaccinia virus assembly. Nature. 1969 Dec 27;224(5226):1280–1284. doi: 10.1038/2241280a0. [DOI] [PubMed] [Google Scholar]
- Moss B., Rosenblum E. N. Letter: Protein cleavage and poxvirus morphogenesis: tryptic peptide analysis of core precursors accumulated by blocking assembly with rifampicin. J Mol Biol. 1973 Dec 5;81(2):267–269. doi: 10.1016/0022-2836(73)90195-2. [DOI] [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983 Jun 25;167(2):391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- Prickett K. S., Amberg D. C., Hopp T. P. A calcium-dependent antibody for identification and purification of recombinant proteins. Biotechniques. 1989 Jun;7(6):580–589. [PubMed] [Google Scholar]
- Rose J. K., Buonocore L., Whitt M. A. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. Biotechniques. 1991 Apr;10(4):520–525. [PubMed] [Google Scholar]
- Rosel J., Moss B. Transcriptional and translational mapping and nucleotide sequence analysis of a vaccinia virus gene encoding the precursor of the major core polypeptide 4b. J Virol. 1985 Dec;56(3):830–838. doi: 10.1128/jvi.56.3.830-838.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov I., Joklik W. K. Studies on the nature and location of the capsid polypeptides of vaccinia virions. Virology. 1972 Nov;50(2):579–592. doi: 10.1016/0042-6822(72)90409-6. [DOI] [PubMed] [Google Scholar]
- Sasaki S., Matsuyama S., Mizushima S. In vitro kinetic analysis of the role of the positive charge at the amino-terminal region of signal peptides in translocation of secretory protein across the cytoplasmic membrane in Escherichia coli. J Biol Chem. 1990 Mar 15;265(8):4358–4363. [PubMed] [Google Scholar]
- Stern W., Pogo B. G., Dales S. Biogenesis of poxviruses: analysis of the morphogenetic sequence using a conditional lethal mutant defective in envelope self-assembly. Proc Natl Acad Sci U S A. 1977 May;74(5):2162–2166. doi: 10.1073/pnas.74.5.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. L., Condit R. C. Marker rescue mapping of vaccinia virus temperature-sensitive mutants using overlapping cosmid clones representing the entire virus genome. Virology. 1986 Apr 15;150(1):10–20. doi: 10.1016/0042-6822(86)90261-8. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanSlyke J. K., Franke C. A., Hruby D. E. Proteolytic maturation of vaccinia virus core proteins: identification of a conserved motif at the N termini of the 4b and 25K virion proteins. J Gen Virol. 1991 Feb;72(Pt 2):411–416. doi: 10.1099/0022-1317-72-2-411. [DOI] [PubMed] [Google Scholar]
- Vanslyke J. K., Whitehead S. S., Wilson E. M., Hruby D. E. The multistep proteolytic maturation pathway utilized by vaccinia virus P4a protein: a degenerate conserved cleavage motif within core proteins. Virology. 1991 Aug;183(2):467–478. doi: 10.1016/0042-6822(91)90976-i. [DOI] [PubMed] [Google Scholar]
- Weber J. Genetic analysis of adenovirus type 2 III. Temperature sensitivity of processing viral proteins. J Virol. 1976 Feb;17(2):462–471. doi: 10.1128/jvi.17.2.462-471.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir J. P., Moss B. Use of a bacterial expression vector to identify the gene encoding a major core protein of vaccinia virus. J Virol. 1985 Nov;56(2):534–540. doi: 10.1128/jvi.56.2.534-540.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellink J., van Kammen A. Proteases involved in the processing of viral polyproteins. Brief review. Arch Virol. 1988;98(1-2):1–26. doi: 10.1007/BF01321002. [DOI] [PubMed] [Google Scholar]
- Yang W. P., Bauer W. R. Purification and characterization of vaccinia virus structural protein VP8. Virology. 1988 Dec;167(2):578–584. [PubMed] [Google Scholar]
- Yang W. P., Kao S. Y., Bauer W. R. Biosynthesis and post-translational cleavage of vaccinia virus structural protein VP8. Virology. 1988 Dec;167(2):585–590. [PubMed] [Google Scholar]
- Zhang Y., Moss B. Immature viral envelope formation is interrupted at the same stage by lac operator-mediated repression of the vaccinia virus D13L gene and by the drug rifampicin. Virology. 1992 Apr;187(2):643–653. doi: 10.1016/0042-6822(92)90467-4. [DOI] [PubMed] [Google Scholar]
- de Groot R. J., Hardy W. R., Shirako Y., Strauss J. H. Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase. Evidence for temporal regulation of polyprotein processing in vivo. EMBO J. 1990 Aug;9(8):2631–2638. doi: 10.1002/j.1460-2075.1990.tb07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]