Abstract
Ras proteins, key regulators of growth, differentiation, and malignant transformation, recently have been implicated in synaptic function and region-specific learning and memory functions in the brain. Rap proteins, members of the Ras small G protein superfamily, can inhibit Ras signaling through the Ras/Raf-1/mitogen-activated protein (MAP) kinase pathway or, through B-Raf, can activate MAP kinase. Rap and Ras proteins both can be activated through guanine nucleotide exchange factors (GEFs). Many Ras GEFs, but to date only one Rap GEF, have been identified. We now report the cloning of a brain-enriched gene, CalDAG-GEFI, which has substrate specificity for Rap1A, dual binding domains for calcium (Ca2+) and diacylglycerol (DAG), and enriched expression in brain basal ganglia pathways and their axon-terminal regions. Expression of CalDAG-GEFI activates Rap1A and inhibits Ras-dependent activation of the Erk/MAP kinase cascade in 293T cells. Ca2+ ionophore and phorbol ester strongly and additively enhance this Rap1A activation. By contrast, CalDAG-GEFII, a second CalDAG-GEF family member that we cloned and found identical to RasGRP [Ebinu, J. O., Bottorff, D. A., Chan, E. Y. W., Stang, S. L., Dunn, R. J. & Stone, J. C. (1998) Science 280, 1082–1088], exhibits a different brain expression pattern and fails to activate Rap1A, but activates H-Ras, R-Ras, and the Erk/MAP kinase cascade under Ca2+ and DAG modulation. We propose that CalDAG-GEF proteins have a critical neuronal function in determining the relative activation of Ras and Rap1 signaling induced by Ca2+ and DAG mobilization. The expression of CalDAG-GEFI and CalDAG-GEFII in hematopoietic organs suggests that such control may have broad significance in Ras/Rap regulation of normal and malignant states.
The basal ganglia are centrally implicated in movement control and in forms of procedural learning related to habit formation (1, 2). It is not yet known whether particular neurochemical specializations of the basal ganglia contribute to these behavioral functions. The basal ganglia do, however, have a unique double-inhibitory pathway design combined with abundant expression of neuromodulators in striatal neurons. A number of genes with differentially high expression in the striatum also have been identified (3–13). These include genes coding for proteins with signaling functions, such as adenylate cyclase V (3) and DARPP-32 (11). In an effort to identify other cellular signaling molecules that could contribute to basal ganglia functions, we initiated a search for striatum-enriched transcripts by a differential display method (14). Among the transcripts identified in this search were a family of genes characterized by the presence of a Ras superfamily guanine nucleotide exchange factor (GEF) domain. Because of the considerable evidence implicating Ras in neuroplasticity and synaptic transmission (15–22), we focused on identifying the functional properties and expression patterns of these genes. We report here on the characterization of CalDAG-GEFI (calcium- and diacylglycerol-regulated GEFI), a striatum-enriched gene product that has binding domains for Ca2+ and DAG, as well as a novel Ras superfamily GEF that targets Rap1 (23–28).
MATERIALS AND METHODS
cDNA Library Screening.
Human full-length CalDAG-GEFI cDNAs were isolated from a human frontal cortex λZAPII cDNA library (Stratagene) and a U937 λZAPII cDNA library (provided by S. Volinia, Università degli Studi di Ferrara, Italy). Mouse full-length CalDAG-GEFI was identified in the mouse expressed sequence tag (EST) database (GenBank accession no. W71787). Rat full-length CalDAG-GEFII cDNA was isolated from a rat whole-brain λZAPII cDNA library (Stratagene) by using human CalDAG-GEFII as a probe. Mouse ESTs identified through blast searches were purchased from Genome Systems (St. Louis).
Northern Analysis of CalDAG-GEFI and CalDag-GEFII mRNA Levels.
Northern blots were purchased from CLONTECH. Probes used included human CalDAG-GEFI, 729-bp EcoRI fragment; human CalDAG-GEFII, 584-bp SacI and HindIII fragment; rat CalDAG-GEFI, 439-bp fragment of EST clone RBC565 (GenBank accession no. C06861), obtained from J. Takeda (Gunma University, Japan); and rat CalDAG-GEFII, 508-bp PCR amplified and subcloned fragment (nucleic acids 2541–3048).
Expression Vectors.
Full-length mouse CalDAG-GEFI cDNA inserted into a pCMV-SPORT expression vector with a carboxyl-terminal FLAG epitope was used for transfection. A PCR-amplified fragment of rat CalDAG-GEFII was subcloned into a pCAGGS expression vector, provided by J. Miazaki (Osaka University, Japan), with the addition of His6-tag at its amino terminus, resulting in pCAGGS-His-CalDAGII. We used pEBG-Krev1 that expresses Rap1A as a fusion protein to glutathione S-transferase (GST) in mammalian cells (28), pEBG-R-Ras, which was obtained from S. Hattori (National Center of Neurobiology and Psychiatry, Japan), other vectors for Ras-family proteins, which were obtained by inserting PCR-amplified cDNAs into pEBG expression vector (28), pCAGGS-C3G and pCAGGS-MSos1 as described previously (28), and pCEV-H-RasV12, which was obtained from K. Kaibuchi (Nara Institute of Science and Technology, Japan).
Analysis of Guanine Nucleotides Bound to Ras-Family Proteins in Intact Cells.
293T cells were transfected by SuperFect (Qiagen) as described previously (28) with expression vectors for GST-tagged Ras-family proteins and with those for various GEFs. Cells were labeled 24 hr after transfection with 32Pi for 2 hr. In some experiments, cells were stimulated with either 10 μM A23187 or 1 μM phorbol 12-myristate 13-acetate (TPA) for 3 min. GST-tagged Ras-family proteins were collected from cell lysates with glutathione Sepharose. Guanine nucleotides bound to Ras-family proteins were separated by TLC.
Activation of Elk1.
Activation of Elk1 was examined by the PathDetect Elk1 transreporting system (Stratagene). 293T cells were transfected with pFR-Luc and pFA-Elk1 with various expression vectors, and light output was detected and analyzed by the use of LAS1000 (Fuji).
Production of Monoclonal and Polyclonal Antibodies Against Mouse CalDAG-GEFI.
His6-tagged mouse CalDAG-GEFI polypeptide (amino acids 349–608) was expressed in bacteria, purified over Ni2+-nitrilotriacetate-agarose (Qiagen), and then used to immunize BALB/c mice. The resultant polyclonal antiserum was monitored by ELISA, Western blot, immunoprecipitation, and immunofluorescence assays on CalDAG-GEFI-transfected COS-7 cells. Hybridomas were generated by PEG (polyethylene glycol)-mediated fusion of donor splenocytes to the SP2/O cell line. Positive hybridoma cell lines were identified by screening in the assays described above and purified by limiting dilution and single-cell cloning. We identified three hybridoma cell lines against mouse CalDAG-GEFI (mAbs 18B11, 2D9, and 18A7) in addition to the polyclonal fusion serum.
Immunohistochemistry.
Lightly postfixed, cryostat-cut, 10-μm thick sections were immunostained by the ABC (Vectastain kit) method for CalDAG-GEFI with mAbs 18B11 and 2D9 and the polyclonal fusion serum, for tyrosine hydroxylase (TH) with mAbs from Incstar (Stillwater, MN), and for μ opioid receptor with polyclonal antiserum from R. P. Elde (University of Minnesota, Minneapolis).
In Situ Hybridization of CalDAG-GEFI to Rat Adult Brain Tissues.
In situ hybridization was performed according to Simmons (29). A 439-bp rat EST clone RBC565 (98.4% identical to mouse CalDAG-GEFI nucleic acids 1777–2216) was isolated by blast search and used for making RNA probes with 33P-labeled UTP (2,000 Ci/mmol, NEN; 1 Ci = 37 GBq) and T3 and T7 RNA polymerase (Promega).
Lesion Studies.
Injections of ibotenic acid (20 μg/μl, 1.5 μl per site, 5-day survival) were made unilaterally at two sites in the midlateral caudoputamen, with contralateral vehicle control injections. Unilateral subthalamic knife-cuts (with control contralateral thalamic knife-cuts) were made at an anteroposterior level between the entopeduncular nucleus and substantia nigra (1- and 3-day survivals). Brains were processed as above for CalDAG-GEFI and TH immunostaining.
RESULTS AND DISCUSSION
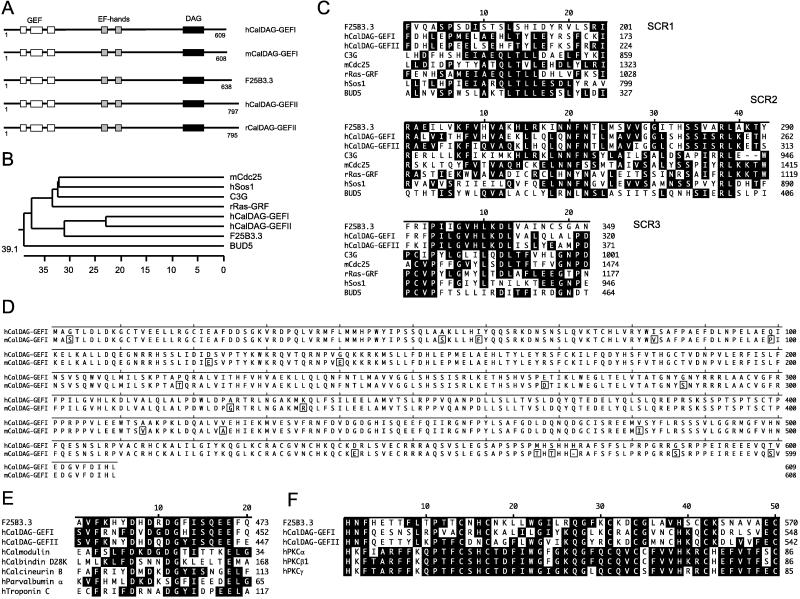
CalDAG-GEFI encodes an approximately 69-kDa protein (Fig. 1D) that displays in its amino-terminal region a GEF domain that is highly homologous to Ras-superfamily GEFs (30) (Fig. 1 A–D). Multiple alignment analysis shows that genes of the CalDAG-GEF family form a cluster within the Ras-GEF superfamily distinct from Ras GEFs such as Sos1 and rRas-GEF (Fig. 1B). The region downstream of the GEF domain contains two tandem repeats of EF-hand Ca2+-binding motifs (31) (Fig. 1 A and E). The carboxyl-terminal region displays a typical DAG/phorbol ester-binding domain, which is present in most PKC family proteins (32) (Fig. 1 A and F).
Figure 1.
(A) CalDAG-GEFs represent a family of Ras-superfamily GEFs including human (h) and mouse (m) CalDAG-GEFI, human (h) and rat (r) CalDAG-GEFII, and Caenorhabditis elegans (F25B3.3, GenBank accession no. 1262950) CalDAG-GEF. (B) Computer-generated phylogenetic tree analysis of the GEF domains of CalDAG-GEFI and CalDAG-GEFII in relation to other Ras-superfamily GEFs. (C) Multiple alignment of GEF structurally conserved regions (SCRs) of CalDAG-GEFs and several other GEFs of the Ras superfamily. (D) Full-length amino acid sequence of human (h) and mouse (m) CalDAG-GEFI (box indicates amino acid differences). (E) Sequence similarity (black indicates identity) of EF-hand domains in CalDAG-GEFs and other calcium-binding proteins. (F) Sequence similarity of DAG-binding domains of CalDAG-GEFs and PKC (protein kinase C) family proteins. Multiple sequence alignments and phylogenetic tree analysis were carried out with the lasergene Software Package (DNAstar, Madison, WI). Abbreviations and GenBank accession nos. of protein sequences are: C3G, 474982; mCdc25, 882120; rRas-GRF, 57665; hSos1 (human son-of-sevenless 1), 476780; BUD5, 171141; hCalmodulin, 115512; hCalbindin D28k, 227666; hCalcineurin B, 105504; hParvalbumin α, 131100; hTroponin C, 136043; hPKCα, 125549; hPKCβ1, 125538; hPKCγ, 462455.
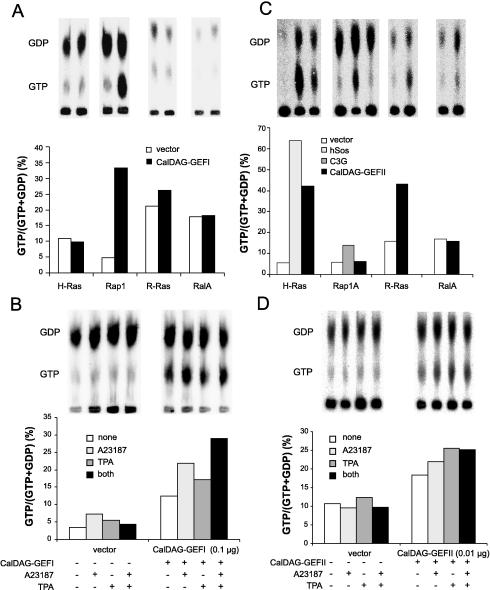
To determine the small G protein target of CalDAG-GEFI, we analyzed guanine nucleotide exchange activity in vivo with intact 293T cells cotransfected with a eukaryotic expression construct of mouse CalDAG-GEFI and GST-tagged Ras-family proteins (28, 33). CalDAG-GEFI transfection produced a dramatic increase in GTP-bound Rap1A compared with control but showed no or minimal activation of H-Ras, R-Ras, or Ral A (Fig. 2A). The increase in GTP-bound Rap1A was augmented in the presence of either the Ca2+ ionophore A23187 or the phorbol ester TPA. Further, A23187 and TPA had additive effects when administered together (Fig. 2B).
Figure 2.
Contrasting functional properties of CalDAG-GEFI and CalDAG-GEFII. (A) Activation of Rap1 by CalDAG-GEFI in 293T cells expressing Ras-family small G proteins with or without CalDAG-GEFI. (B) Calcium ionophore and TPA enhancement of Rap1A activation by CalDAG-GEFI in 293T cells. (C) CalDAG-GEFII activates H-Ras and R-Ras in intact 293T cells. GST-tagged H-Ras, Rap1A, R-Ras, or RalA were expressed in 293T cells with or without Sos1, C3G, or CalDAG-GEFII. (D) Augmentation of H-Ras activation by CalDAG-GEFII in the presence of A23187 or TPA or both, assayed as in B.
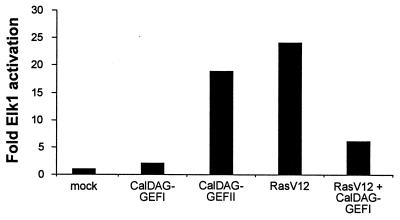
To determine the effect of CalDAG-GEFI on the Erk/MAP kinase cascade, we measured Elk1 activation in 293T cells transfected with CalDAG-GEFI or constitutively active H-Ras (RasV12), or both. CalDAG-GEFI reduced RasV12 activation of Elk1 by ≈4-fold and did not itself activate Elk1 (Fig. 3). Thus, CalDAG-GEFI strongly inhibits Ras-dependent stimulation of the Erk/MAP kinase cascade.
Figure 3.
Inhibition of Ras/Erk/Elk1 signaling cascade by CalDAG-GEFI and activation of this cascade by CalDAG-GEFII. 293T cells were transfected with pFR-Luc and pFA-Elk1 together with expression vectors as indicated.
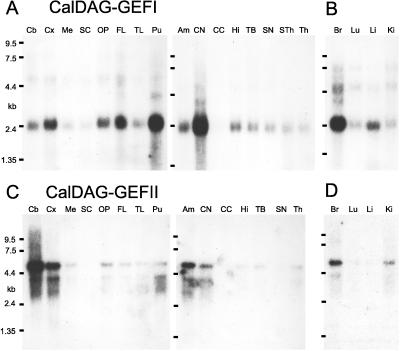
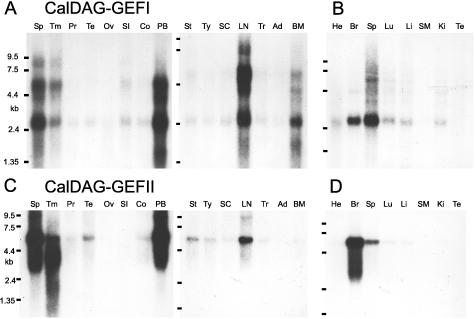
Northern analysis showed that human CalDAG-GEFI is expressed strongly in the brain and that CalDAG-GEFI mRNA is strikingly enriched in the striatum (Fig. 4A). Weaker hybridization was detectable elsewhere. In situ hybridization of sections from the adult rat brain confirmed these restricted distribution patterns (Fig. 5 A and A′). Intense signal was present in the striatum (caudoputamen) and the ventral striatum (nucleus accumbens, olfactory tubercle). There was a weaker signal in the olfactory bulb and in some other sites (Fig. 5A).
Figure 4.
Northern hybridization analysis of CalDAG-GEFI and CalDAG-GEFII in adult human brain. (A) Enriched expression of CalDAG-GEFI in the putamen and caudate nucleus. Compare with C, showing that human brain CalDAG-GEFII mRNA expression levels are most enriched in cerebellum, cerebral cortex, and amygdala. B and D show high brain expression in human fetus. Am, amygdala; Br, brain; Cb, cerebellum; CC, corpus callosum; CN, caudate nucleus; Cx, cortex; FL, frontal lobe; Hi, hippocampus; Ki, kidney; Li, liver; Lu, lung; Me, medulla oblongata; OP, occipital pole; Pu, putamen; SC, spinal cord; SN, substantia nigra; Sth, subthalamic nucleus; TB, total brain; Th, thalamus; TL, temporal lobe.
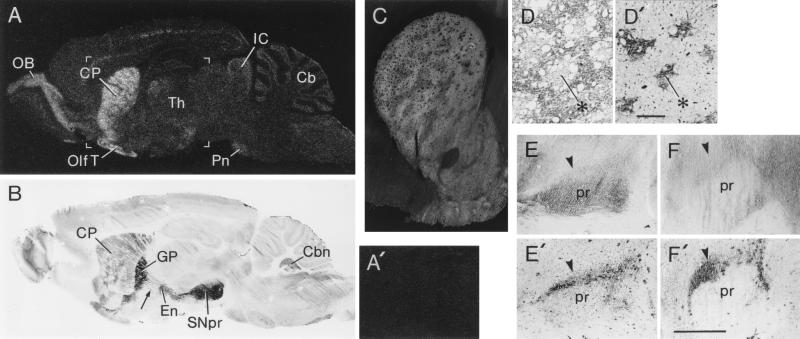
Figure 5.
Preferential distribution of CalDAG-GEFI in the basal ganglia. (A) In situ hybridization film autoradiogram of parasagittal section through rat brain demonstrating predominant localization of CalDAG-GEFI mRNA in the striatum and olfactory bulb with weaker signal in other brain sites. (A′) Sense hybridization in control section shows no label in striatum. Area shown roughly corresponds to bracketed region in A. (B) Immunohistochemical staining of parasagittal section through rat brain with CalDAG-GEFI mAb 18B11 shows strong staining of striatum and striatal projection fibers, and intense staining of globus pallidus and substantia nigra, with significant staining also in olfactory bulb and brainstem (including cerebellar) nuclei. Arrow points to immunostained striatal projection fibers. (C) Reverse-contrast photomicrograph of CalDAG-GEFI immunoreactivity (which appears light) in rat striatum, demonstrating predominant expression of CalDAG-GEFI in the striatal matrix compartment. (D and D′) Correspondence of CalDAG-GEFI-poor zones (D) in rat caudoputamen with μ-opioid rich striosomes (D′, example at asterisks) stained in adjoining sections. (Bar = 200 μm.) (E–F′) Knife-cut through subthalamic region sharply decreases CalDAG-GEFI immunostaining in substantia nigra. Anterior to left. Arrowheads point to substantia nigra pars compacta. (E) Control side (with thalamic knife-cut sparing striatal projection fibers) shows strong CalDAG-GEFI immunostaining. (E′) Serially adjoining section stained for TH showing location of TH-positive substantia nigra pars compacta. (F) Reduced CalDAG-GEFI immunostaining in substantia nigra on side of subthalamic knife-cut. (F′) Serially adjoining section showing TH-positive substantia nigra pars compacta. (Bar = 1 mm.) Cb, cerebellum; Cbn, cerebellar nuclei; CP, caudoputamen; En, entopeduncular nucleus; GP, globus pallidus; IC, inferior colliculus; OB, olfactory bulb; Olf T, olfactory tubercle; Pn, pontine nuclei; pr, substantia nigra pars reticulata; SNpr, substantia nigra pars reticulata; Th, thalamus.
A series of mAbs against the carboxyl-terminal half of mouse CalDAG-GEFI was raised. Western analysis showed that mAbs 18B11 and 2D9 were specific for CalDAG-GEFI. Immunohistochemistry with mAb 18B11 showed a striking basal ganglia-enriched distribution pattern in sections of adult rat brain, with significant but weaker activity elsewhere (Fig. 5 B–D′). CalDAG-GEFI immunoreactivity marked the entire pathway from the striatal matrix compartment to the pallidum and substantia nigra pars reticulata, where very intense CalDAG-GEFI staining was present (Fig. 5B). This pattern, taken together with the in situ hybridization data, strongly suggests that CalDAG-GEFI is synthesized in striatal projection neurons and is transported to striatopallidal and striatonigral terminals.
We tested this possibility in rats by making unilateral intrastriatal injections of ibotenic acid and, in other rats, by making subthalamic knife-cuts to sever striatonigral efferents. These procedures all reduced CalDAG-GEFI staining in the substantia nigra (Fig. 5 E–F′), indicating that CalDAG-GEFI is a protein transported in striatal axons to their terminals. The terminal localization of CalDAG-GEFI was confirmed in subcellular fractionation experiments on dissected samples from the rat ventral midbrain, in which Western analysis showed the presence of CalDAG-GEFI in cytosol and in membrane fractions, including synaptosomes (data not shown).
Because of the similarity of the GEF domains of CalDAG-GEFI and CalDAG-GEFII (Fig. 1C), we examined the substrate specificity of CalDAG-GEFII with the same 293T cell assay system used for CalDAG-GEFI. We confirmed that CalDAG-GEFII can activate Ras (34), and further showed that it can activate H-Ras and R-Ras, but not Ral A or Rap1A (Fig. 2C). H-Ras activation was enhanced by A23187 and TPA (Fig. 2D). Moreover, CalDAG-GEFII, unlike CalDAG-GEFI, increased the transcriptional activity of Elk1 downstream to Erk/MAP kinase (Fig. 3). Thus, in the 293T system, CalDAG-GEFI and CalDag-GEFII target different Ras-superfamily small G proteins and have opposite effects on the MAP kinase cascade. Northern analysis further showed contrasting brain expression for CalDAG-GEFII, with highest expression being in the cerebellum, cerebral cortex, and amygdala, and low expression occurring in the striatum (Fig. 4C). Interestingly, both genes also are expressed in hematopoietic organs in both human and rat (Fig. 6A–D).
Figure 6.
CalDAG-GEFI and CalDAG-GEFII are expressed in the hematopoietic system. (A and B) CalDAG-GEFI expression in adult human (A) and rat (B) organs. (C and D) CalDAG-GEFII expression in adult human (C) and rat (D) organs. Ad, adrenal gland; BM, bone marrow; Br, brain; Co, colon (mucosal lining); He, heart; Ki, kidney; Li, liver; LN, lymph node; Lu, lung; Ov, ovary; PB, peripheral blood leukocytes; Pr, prostate; SC, spinal cord; SI, small intestine; SM, skeletal muscle; Sp, spleen; St, stomach; Te, testis; Tm, thymus; Tr, trachea; Ty, thyroid.
Our findings have significance of two sorts. First, they suggest that Rap signaling may be important in regulating basal ganglia output in response to Ca2+ and DAG. It is known that corticostriatal inputs can activate the MAP kinase cascade in striatal projection neurons (35) and that phosphoinositide (PI) signaling is represented strongly in these pathways (36). Moreover, a number of receptor systems in the striatum and its striatonigral/striatopallidal pathways are linked to Ca2+ and PI signaling, notably including N-methyl-d-aspartate and metabotropic glutamate receptors, D2-class dopamine receptors, and tachykinin receptors (37, 38). Our experiments suggest that a previously unrecognized signaling target for some of these systems is likely to be Rap1, via CalDAG-GEFI. Our evidence for heavy accumulation of CalDAG-GEFI in the target nuclei of striatal outputs raises the possibility that CalDAG-GEFI has a synaptic function. The reported localization of Rap1 in synaptosomes and synaptic vesicles (39) supports this view. The particular basal ganglia projection systems that we have identified as enriched in CalDAG-GEFI are differentially vulnerable to neurodegeneration in Huntington’s disease. Rap/Ras function in these pathways should be considered in analyzing this vulnerability and growth factor-based therapeutic strategies (40).
A second and more general implication of our findings is that Rap and Ras functions can be regulated coordinately or disjunctively by Ca2+ and DAG in the brain and hematopoietic organs, depending on the relative expression of CalDAG-GEFI and CalDAG-GEFII. In neurons, Ras/MAP kinase signaling has been implicated directly in synaptic transmission and the neuroplasticity underlying learning and memory (15–22). Different CalDAG-GEFI and CalDAG-GEFII expression patterns in the brain may critically influence region-specific neuroplasticity mediated by Ca2+- and DAG-signaling pathways. The presence of CalDAG-GEFI and CalDAG-GEFII in the hematopoietic system further suggests a direct input of Ca2+ and DAG to Ras/Rap regulation of normal growth and differentiation as well as malignant transformation.
Acknowledgments
We thank Dr. J. Lees and Dr. K. Moberg for their help with antibody production, Mr. H. F. Hall and Mr. G. Holm for their help with the illustrations, and Drs. S. Volinia, R. P. Elde, S. Hattori, J. Miyazaki, K. Kaibuchi, and J. Takeda for reagents. This work was funded by the generous support of the Grayce B. Kerr Fund, the James and Pat Poitras Research Fund, and the Stanley Foundation, by grants from the National Institutes of Health (NICHD R01 HD28341, NCI P01 CA42063, NHLBI P01 HL41484, and NCHGR R01 HG00299), and by the Japan Science and Technology Agency and Health Sciences Foundation.
ABBREVIATIONS
- CalDAG-GEFI or CalDag-GEFII
calcium- and diacylglycerol-regulated guanine nucleotide exchange factor I or II, respectively
- DAG
diacylglycerol
- MAP
mitogen-activated protein
- GEF
guanine nucleotide exchange factor
- TH
tyrosine hydroxylase
- GST
glutathione S-transferase
- TPA
phorbol 12-myristate 13-acetate
Footnotes
References
- 1. Graybiel A M. Neurobiol Learning Memory. 1998;70:119–136. doi: 10.1006/nlme.1998.3843. [DOI] [PubMed] [Google Scholar]
- 2.Knowlton B J, Mangels J A, Squire L R. Science. 1996;273:1399–1402. doi: 10.1126/science.273.5280.1399. [DOI] [PubMed] [Google Scholar]
- 3.Glatt C E, Snyder S H. Nature (London) 1993;361:536–538. doi: 10.1038/361536a0. [DOI] [PubMed] [Google Scholar]
- 4.Drinnan S L, Hope B T, Snutch T P, Vincent S R. Mol Cell Neurosci. 1991;2:66–70. doi: 10.1016/1044-7431(91)90040-u. [DOI] [PubMed] [Google Scholar]
- 5.Celio M R. Neuroscience. 1990;35:375–475. doi: 10.1016/0306-4522(90)90091-h. [DOI] [PubMed] [Google Scholar]
- 6.Ouimet C C, Hemmings H C, Jr, Greengard P. J Neurosci. 1989;9:865–875. doi: 10.1523/JNEUROSCI.09-03-00865.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan C, Bentley J K, Sonnenburg W K, Beavo J A. J Neurosci. 1994;14:973–984. doi: 10.1523/JNEUROSCI.14-03-00973.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lombroso P J, Murdoch G, Lerner M. Proc Natl Acad Sci USA. 1991;88:7242–7246. doi: 10.1073/pnas.88.16.7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watson J B, Coulter P M, Sutcliffe J G. Brain Dysfunction. 1992;5:94–105. [Google Scholar]
- 10.Usui H, Falk J D, Dopaza A, de Lecea L, Erlander M G, Sutcliffe J G. J Neurosci. 1994;14:4915–4926. doi: 10.1523/JNEUROSCI.14-08-04915.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemmings H C, Jr, Greengard P, Tung H Y L, Cohen P. Nature (London) 1984;310:502–505. doi: 10.1038/310503a0. [DOI] [PubMed] [Google Scholar]
- 12.Berhow M T, Hiroi N, Nestler E J. J Neurosci. 1996;16:4707–4715. doi: 10.1523/JNEUROSCI.16-15-04707.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ortiz J, Harris H W, Guitart X, Terwilliger R Z, Haycock J W, Nestler E J. J Neurosci. 1995;15:1285–1297. doi: 10.1523/JNEUROSCI.15-02-01285.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang P, Pardee A B. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 15.Silva A J, Frankland P W, Marowitz Z, Friedman E, Lazlo G, Cioffi D, Jacks T, Bourtchuladze R. Nat Genet. 1997;15:281–284. doi: 10.1038/ng0397-281. [DOI] [PubMed] [Google Scholar]
- 16.Sturani E, Abbondio A, Branduardi P, Ferrari C, Zippel R, Martegani E, Vanoni M, Denis-Donini S. Exp Cell Res. 1997;235:117–123. doi: 10.1006/excr.1997.3660. [DOI] [PubMed] [Google Scholar]
- 17.Zippel R, Gnesutta N, Matus-Leibovitch N, Mancinelli E, Saya D, Vogel Z, Sturani E, Renata Z, Nerina G, Noa M L, et al. Brain Res Mol Brain Res. 1997;48:140–144. doi: 10.1016/s0169-328x(97)00120-4. [DOI] [PubMed] [Google Scholar]
- 18.Brambilla R, Gnesutta N, Minichiello L, White G, Roylance A J, Herron C E, Ramsey M, Wolfer D P, Cestari V, Rossi-Arnaud C, et al. Nature (London) 1997;390:281–285. doi: 10.1038/36849. [DOI] [PubMed] [Google Scholar]
- 19.Farnsworth C L, Freshney N W, Rosen L B, Ghosh A, Greenberg M E, Feig L A. Nature (London) 1995;376:524–527. doi: 10.1038/376524a0. [DOI] [PubMed] [Google Scholar]
- 20.Rosen L B, Ginty D D, Weber M J, Greenberg M E. Neuron. 1994;12:1207–1221. doi: 10.1016/0896-6273(94)90438-3. [DOI] [PubMed] [Google Scholar]
- 21.Chen H-J, Rojas-Soto M, Oguni A, Kennedy M B. Neuron. 1998;20:895–904. doi: 10.1016/s0896-6273(00)80471-7. [DOI] [PubMed] [Google Scholar]
- 22.Kim J H, Liao D, Lau L-F, Huganir R L. Neuron. 1998;20:683–691. doi: 10.1016/s0896-6273(00)81008-9. [DOI] [PubMed] [Google Scholar]
- 23.Bokoch G M. Biochem J. 1993;289:17–24. doi: 10.1042/bj2890017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohtsuka T, Shimizu K, Yamamori B, Kuroda S, Takai Y. J Biol Chem. 1996;271:1258–1261. doi: 10.1074/jbc.271.3.1258. [DOI] [PubMed] [Google Scholar]
- 25.Burgering B M T, Bos J L. Trends Biochem Sci. 1995;20:18–22. doi: 10.1016/s0968-0004(00)88944-6. [DOI] [PubMed] [Google Scholar]
- 26.York R D, Yao H, Dillon T, Ellig C L, Eckert S P, McCleskey E W, Stork P J. Nature (London) 1998;392:622–626. doi: 10.1038/33451. [DOI] [PubMed] [Google Scholar]
- 27.Vossler M R, Yao H, York R D, Pan M G, Rim C S, Stork P J. Cell. 1997;89:73–82. doi: 10.1016/s0092-8674(00)80184-1. [DOI] [PubMed] [Google Scholar]
- 28.Gotoh T, Hattori S, Nakamura S, Kitayama H, Noda M, Takai Y, Kaibuchi K, Matsui H, Hatase O, Takahashi H, et al. Mol Cell Biol. 1995;15:6746–6753. doi: 10.1128/mcb.15.12.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simmons D, Arriza J, Swanson L. J Histotechnol. 1989;12:169–181. [Google Scholar]
- 30.Boguski M S, McCormick F. Nature (London) 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 31.Heizmann C W. Gen Physiol Biophys. 1992;11:411–425. [PubMed] [Google Scholar]
- 32.Newton A C. J Biol Chem. 1995;270:28495–28498. doi: 10.1074/jbc.270.48.28495. [DOI] [PubMed] [Google Scholar]
- 33.Franke B, Akkerman J W, Bos J L. EMBO J. 1997;16:252–259. doi: 10.1093/emboj/16.2.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ebinu J O, Bottorff D A, Chan E Y W, Stang S L, Dunn R J, Stone J C. Science. 1998;280:1082–1088. doi: 10.1126/science.280.5366.1082. [DOI] [PubMed] [Google Scholar]
- 35.Sgambato V, Vanhoutte P, Pages C, Rogard M, Hipskind R, Besson M J, Caboche J. J Neurosci. 1998;18:214–226. doi: 10.1523/JNEUROSCI.18-01-00214.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fotuhi M, Dawson T M, Sharp A H, Martin L J, Graybiel A M, Snyder S H. J Neurosci. 1993;13:3300–3308. doi: 10.1523/JNEUROSCI.13-08-03300.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graybiel A M, Penney J B. In: Handbook of Chemical Neuroanatomy. Bloom F E, editor. Vol. 13. Amsterdam: Elsevier; 1998. , in press. [Google Scholar]
- 38.Fiorillo C D, Williams J T. Nature (London) 1998;394:78–82. doi: 10.1038/27919. [DOI] [PubMed] [Google Scholar]
- 39.Kim S, Mizoguchi A, Kikuchi A, Takai Y. Mol Cell Biol. 1990;10:2645–2652. doi: 10.1128/mcb.10.6.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson K D, Panayotatos N, Corcoran T L, Lindsay R M, Wiegand S J. Proc Natl Acad Sci USA. 1996;93:7346–7351. doi: 10.1073/pnas.93.14.7346. [DOI] [PMC free article] [PubMed] [Google Scholar]