Abstract
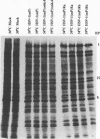
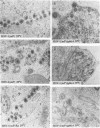
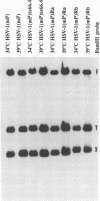
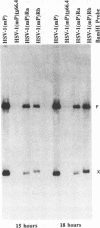
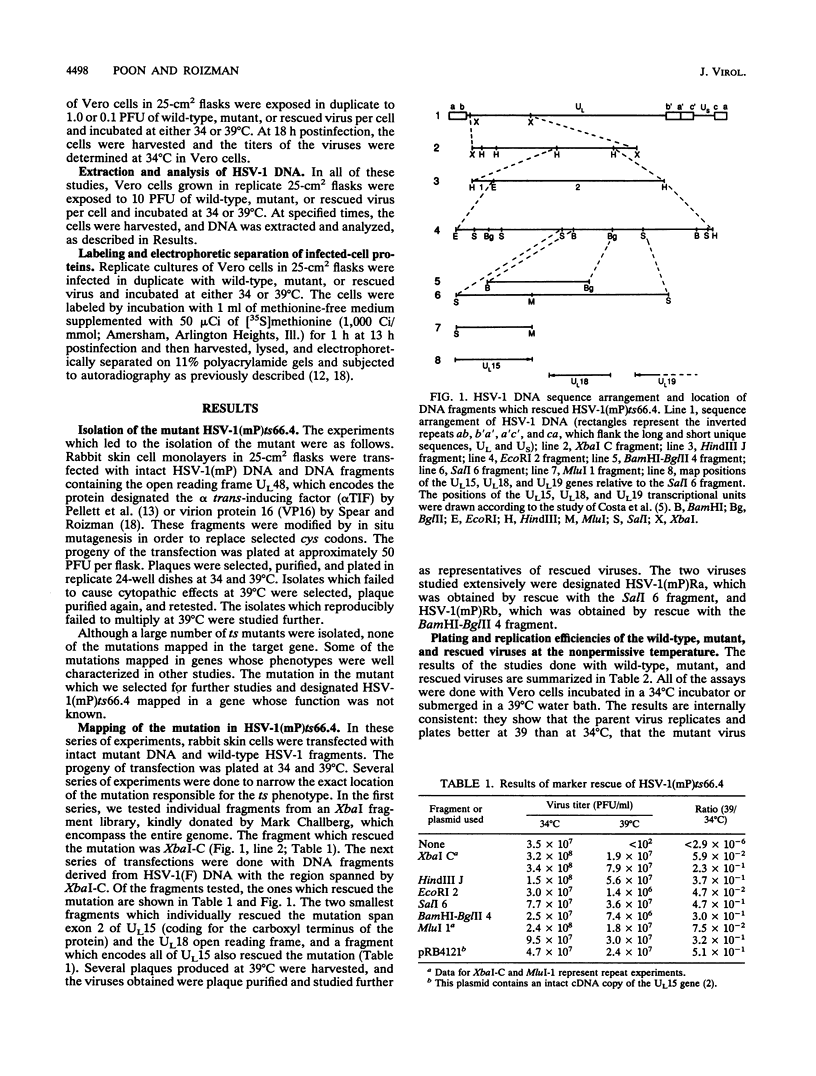
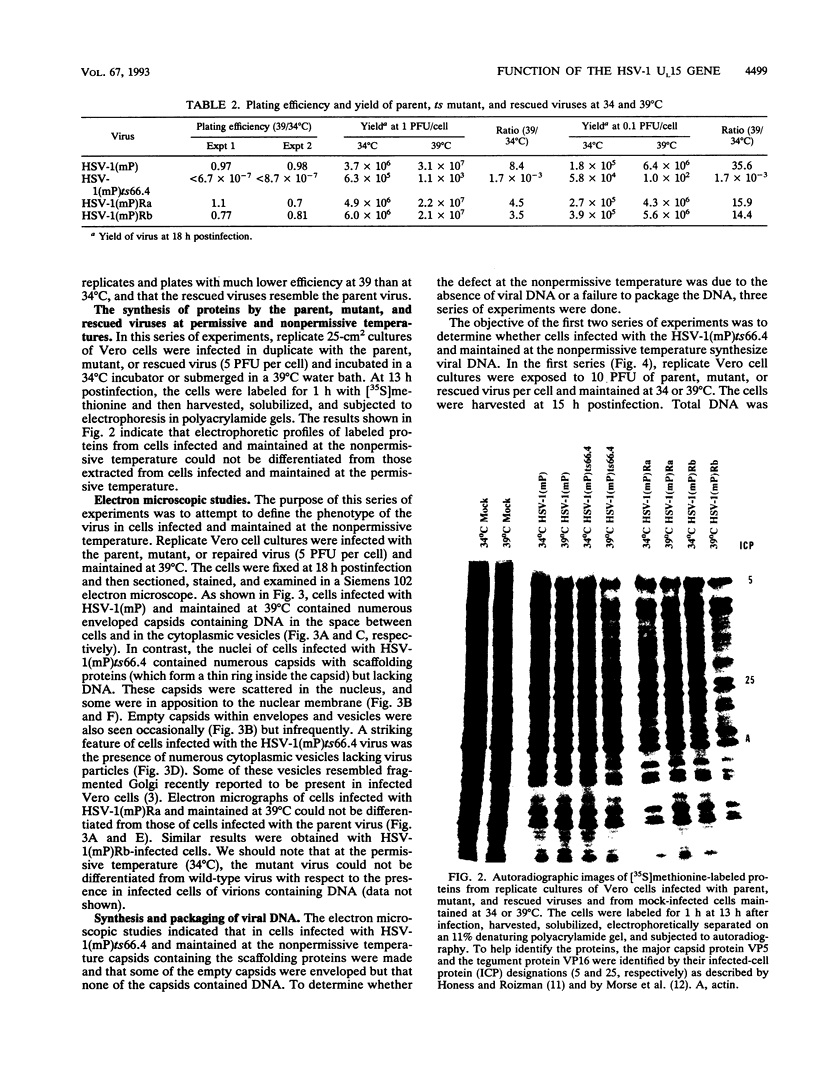
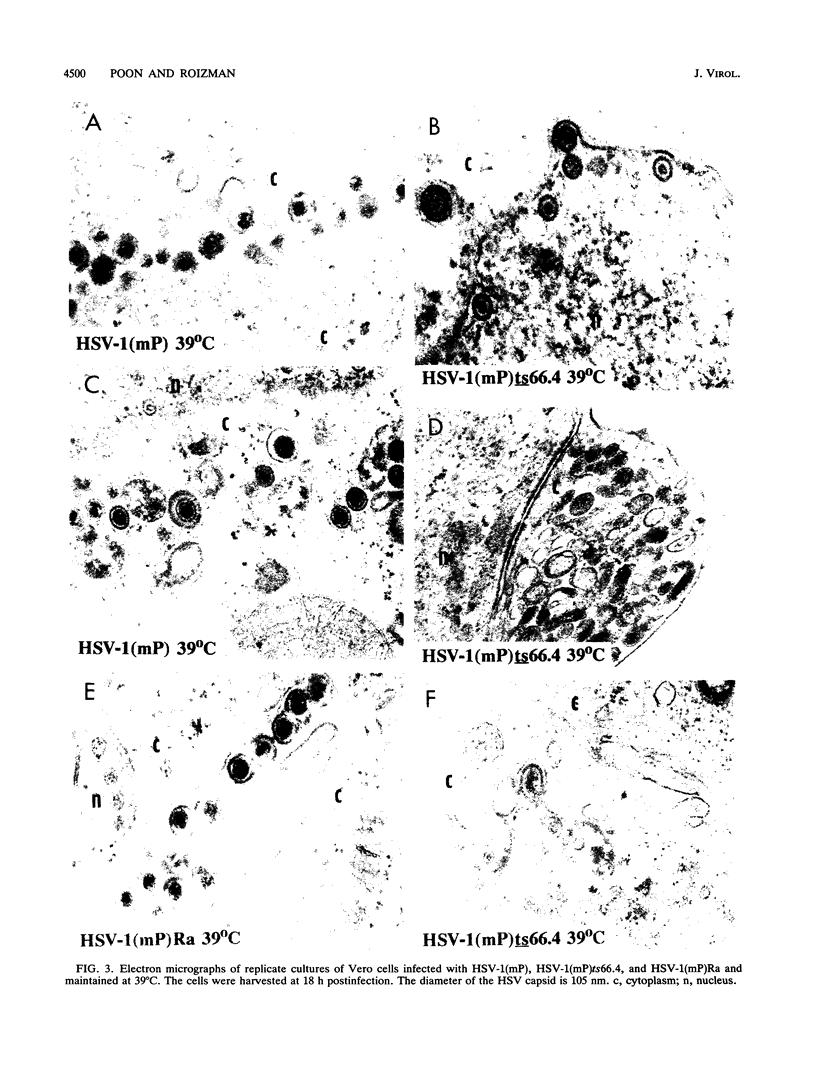
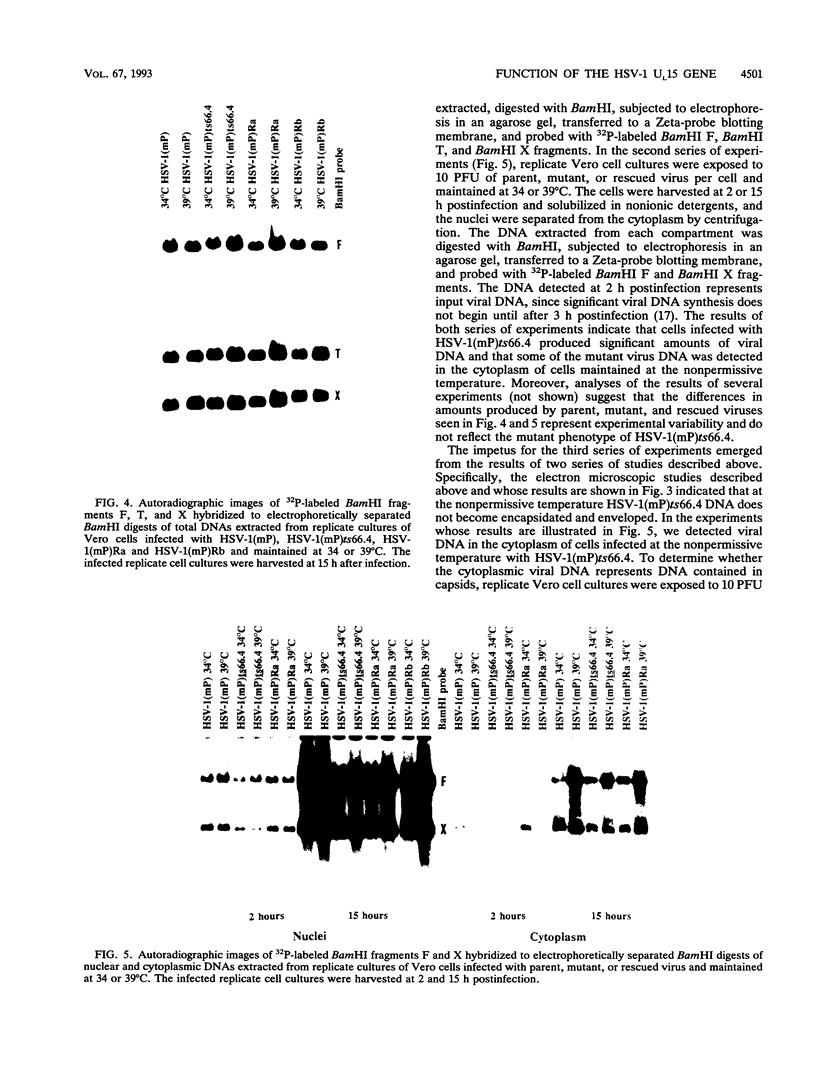
The UL15 gene of herpes simplex virus 1 consists of two exons and is highly conserved among the herpesviruses sequenced to date. Other than its homology to a phage protein involved in the packaging of DNA, nothing is known of its function. This report concerns the isolation of a temperature-sensitive mutant with a mutation mapping in the UL15 open reading frame. Cells infected with the parent, mutant, and rescued viruses all make DNA at the nonpermissive temperature. Direct analyses of the DNA and electron microscopic studies indicate that although viral DNA is made, it is not packaged into capsids present in nuclei. These studies suggest that UL15 may be involved in the packaging of viral DNA.
Full text
PDF
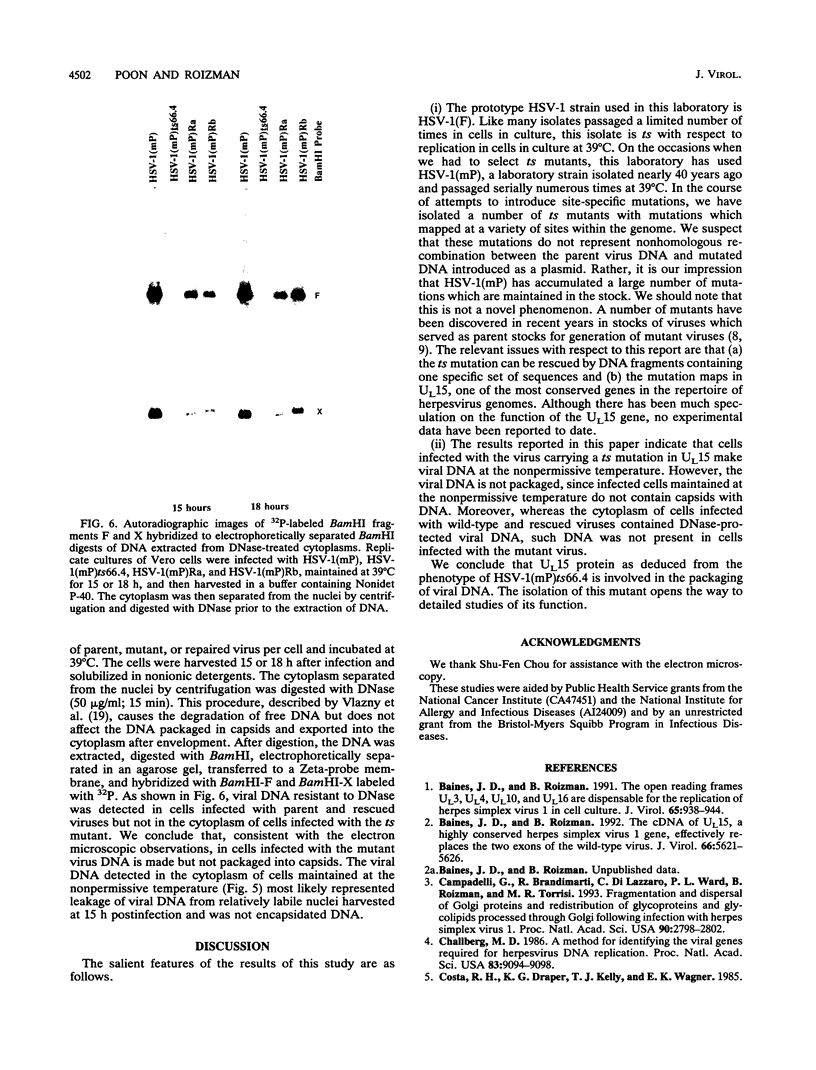

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baines J. D., Roizman B. The cDNA of UL15, a highly conserved herpes simplex virus 1 gene, effectively replaces the two exons of the wild-type virus. J Virol. 1992 Sep;66(9):5621–5626. doi: 10.1128/jvi.66.9.5621-5626.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines J. D., Roizman B. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J Virol. 1991 Feb;65(2):938–944. doi: 10.1128/jvi.65.2.938-944.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campadelli G., Brandimarti R., Di Lazzaro C., Ward P. L., Roizman B., Torrisi M. R. Fragmentation and dispersal of Golgi proteins and redistribution of glycoproteins and glycolipids processed through the Golgi apparatus after infection with herpes simplex virus 1. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2798–2802. doi: 10.1073/pnas.90.7.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challberg M. D. A method for identifying the viral genes required for herpesvirus DNA replication. Proc Natl Acad Sci U S A. 1986 Dec;83(23):9094–9098. doi: 10.1073/pnas.83.23.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R. H., Draper K. G., Kelly T. J., Wagner E. K. An unusual spliced herpes simplex virus type 1 transcript with sequence homology to Epstein-Barr virus DNA. J Virol. 1985 May;54(2):317–328. doi: 10.1128/jvi.54.2.317-328.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A. J. Channel catfish virus: a new type of herpesvirus. Virology. 1992 Jan;186(1):9–14. doi: 10.1016/0042-6822(92)90056-u. [DOI] [PubMed] [Google Scholar]
- Ejercito P. M., Kieff E. D., Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968 May;2(3):357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- Everett R. D., Fenwick M. L. Comparative DNA sequence analysis of the host shutoff genes of different strains of herpes simplex virus: type 2 strain HG52 encodes a truncated UL41 product. J Gen Virol. 1990 Jun;71(Pt 6):1387–1390. doi: 10.1099/0022-1317-71-6-1387. [DOI] [PubMed] [Google Scholar]
- Fisher F. B., Preston V. G. Isolation and characterisation of herpes simplex virus type 1 mutants which fail to induce dUTPase activity. Virology. 1986 Jan 15;148(1):190–197. doi: 10.1016/0042-6822(86)90414-9. [DOI] [PubMed] [Google Scholar]
- HOGGAN M. D., ROIZMAN B. The isolation and properties of a variant of Herpes simplex producing multinucleated giant cells in monolayer cultures in the presence of antibody. Am J Hyg. 1959 Sep;70:208–219. doi: 10.1093/oxfordjournals.aje.a120071. [DOI] [PubMed] [Google Scholar]
- Honess R. W., Roizman B. Proteins specified by herpes simplex virus. XI. Identification and relative molar rates of synthesis of structural and nonstructural herpes virus polypeptides in the infected cell. J Virol. 1973 Dec;12(6):1347–1365. doi: 10.1128/jvi.12.6.1347-1365.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse L. S., Pereira L., Roizman B., Schaffer P. A. Anatomy of herpes simplex virus (HSV) DNA. X. Mapping of viral genes by analysis of polypeptides and functions specified by HSV-1 X HSV-2 recombinants. J Virol. 1978 May;26(2):389–410. doi: 10.1128/jvi.26.2.389-410.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellett P. E., McKnight J. L., Jenkins F. J., Roizman B. Nucleotide sequence and predicted amino acid sequence of a protein encoded in a small herpes simplex virus DNA fragment capable of trans-inducing alpha genes. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5870–5874. doi: 10.1073/pnas.82.17.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post L. E., Conley A. J., Mocarski E. S., Roizman B. Cloning of reiterated and nonreiterated herpes simplex virus 1 sequences as BamHI fragments. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4201–4205. doi: 10.1073/pnas.77.7.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell D., Franklin J., Arisaka F., Mosig G. Bacteriophage T4 DNA packaging genes 16 and 17. Nucleic Acids Res. 1990 Jul 11;18(13):4005–4005. doi: 10.1093/nar/18.13.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROIZMAN B., AURELIAN L., ROANE P. R., Jr THE MULTIPLICATION OF HERPES SIMPLEX VIRUS. I. THE PROGRAMMING OF VIRAL DNA DUPLICATION IN HEP-2 CELLS. Virology. 1963 Nov;21:482–498. doi: 10.1016/0042-6822(63)90209-5. [DOI] [PubMed] [Google Scholar]
- Rao V. B., Black L. W. Cloning, overexpression and purification of the terminase proteins gp16 and gp17 of bacteriophage T4. Construction of a defined in-vitro DNA packaging system using purified terminase proteins. J Mol Biol. 1988 Apr 5;200(3):475–488. doi: 10.1016/0022-2836(88)90537-2. [DOI] [PubMed] [Google Scholar]
- Spear P. G., Roizman B. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J Virol. 1972 Jan;9(1):143–159. doi: 10.1128/jvi.9.1.143-159.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlazny D. A., Kwong A., Frenkel N. Site-specific cleavage/packaging of herpes simplex virus DNA and the selective maturation of nucleocapsids containing full-length viral DNA. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1423–1427. doi: 10.1073/pnas.79.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]