Abstract
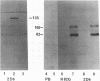
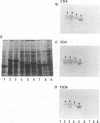
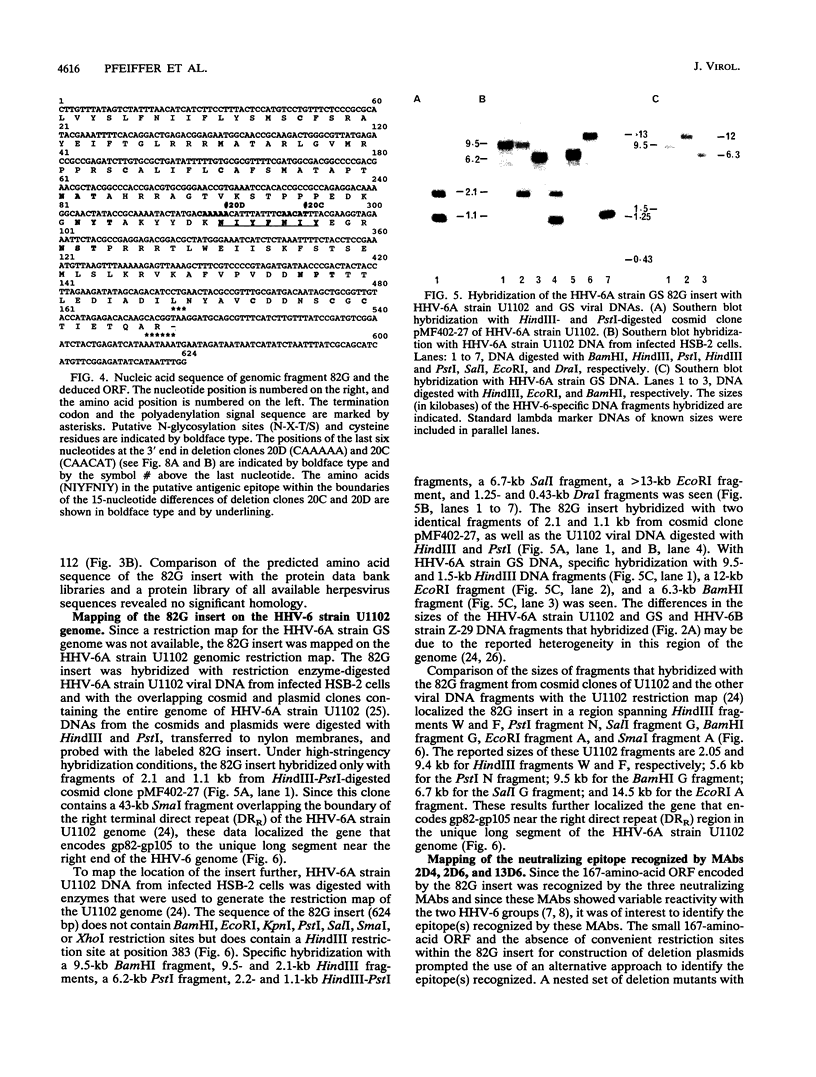
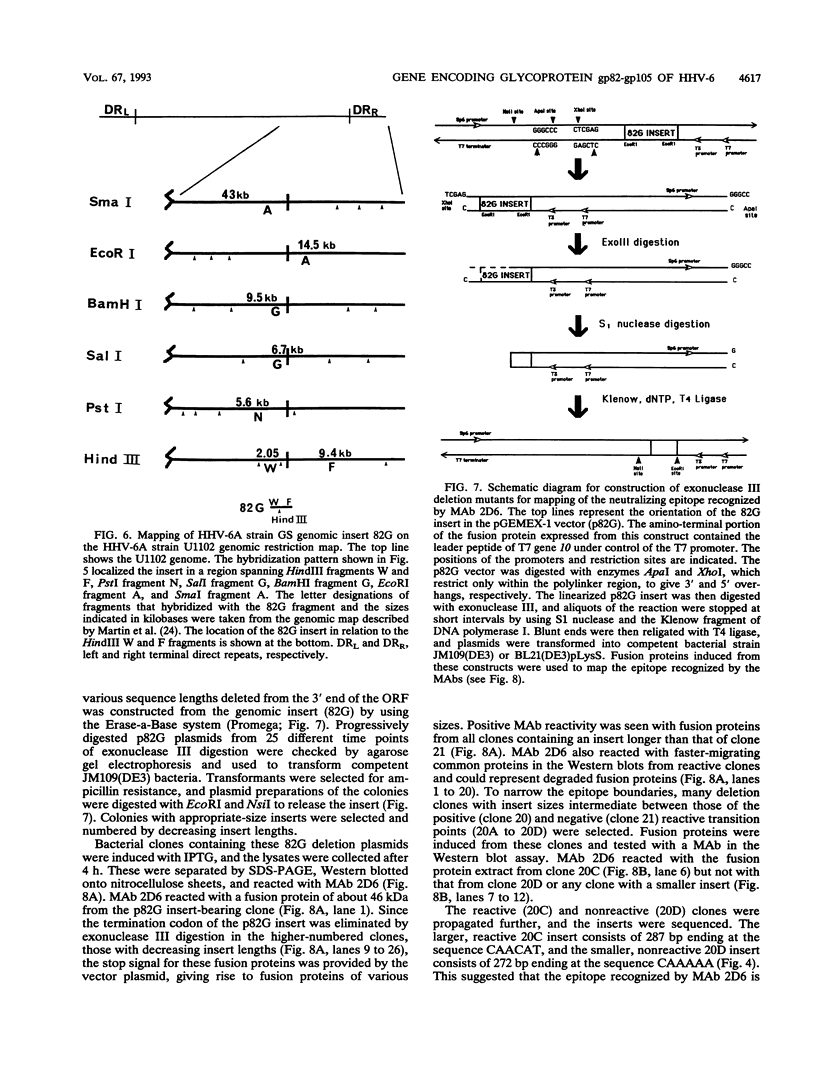
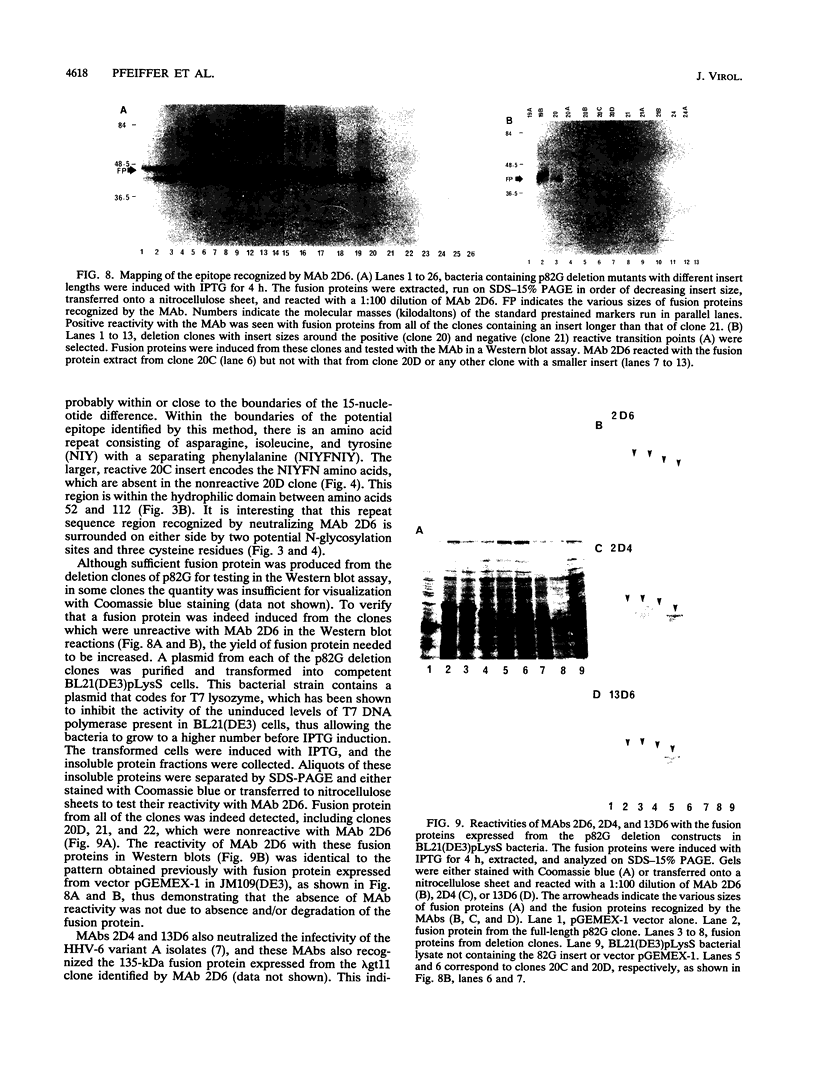
Monoclonal antibodies (MAbs) 2D4, 2D6, and 13D6 against human herpesvirus 6 (HHV-6) variant A strain GS recognized virion envelope glycoprotein complex gp82-gp105 and neutralized the infectivity of HHV-6 variant A group isolates. A 624-bp genomic fragment (82G) was identified from an HHV-6 strain GS genomic library constructed in the lambda gt11 expression system by immunoscreening with MAb 2D6. Rabbit antibodies against the fusion protein expressed from the genomic insert recognized glycoprotein complex gp82-gp105 from HHV-6-infected cells, thus confirming that the genomic fragment is a portion of the gene(s) that encodes gp82-gp105. This genomic insert hybridized specifically with viral DNAs from HHV-6 variant A strains GS and U1101 under high-stringency conditions but hybridized with HHV-6 variant B strain Z-29 DNA only under low-stringency conditions. DNA sequence analysis of the insert revealed a 167-amino-acid single open reading frame with an open 5' end and a stop codon at the 3' end. Hybridization studies with HHV-6A strain U1102 DNA localized the gp82-gp105-encoding gene to the unique long region near the direct repeat at the right end of the genome. To locate the neutralizing epitope(s) recognized by the MAbs, a series of deletions from the 3' end of the gene were constructed with exonuclease III, and fusion proteins from deletion constructs were tested for reactivity with MAbs in a Western immunoblot assay. Sequencing of deletion constructs at the reactive-nonreactive transition point localized the epitope recognized by the three neutralizing MAbs within or near a repeat amino acid sequence (NIYFNIY) of the putative protein. This repeat sequence region is surrounded on either side by two potential N-glycosylation sites and three cysteine residues.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ablashi D. V., Balachandran N., Josephs S. F., Hung C. L., Krueger G. R., Kramarsky B., Salahuddin S. Z., Gallo R. C. Genomic polymorphism, growth properties, and immunologic variations in human herpesvirus-6 isolates. Virology. 1991 Oct;184(2):545–552. doi: 10.1016/0042-6822(91)90424-a. [DOI] [PubMed] [Google Scholar]
- Ablashi D. V., Josephs S. F., Buchbinder A., Hellman K., Nakamura S., Llana T., Lusso P., Kaplan M., Dahlberg J., Memon S. Human B-lymphotropic virus (human herpesvirus-6). J Virol Methods. 1988 Sep;21(1-4):29–48. doi: 10.1016/0166-0934(88)90050-x. [DOI] [PubMed] [Google Scholar]
- Aubin J. T., Collandre H., Candotti D., Ingrand D., Rouzioux C., Burgard M., Richard S., Huraux J. M., Agut H. Several groups among human herpesvirus 6 strains can be distinguished by Southern blotting and polymerase chain reaction. J Clin Microbiol. 1991 Feb;29(2):367–372. doi: 10.1128/jcm.29.2.367-372.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran N., Amelse R. E., Zhou W. W., Chang C. K. Identification of proteins specific for human herpesvirus 6-infected human T cells. J Virol. 1989 Jun;63(6):2835–2840. doi: 10.1128/jvi.63.6.2835-2840.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran B., Tirawatnapong S., Pfeiffer B., Ablashi D. V. Antigenic relationships among human herpesvirus-6 isolates. J Med Virol. 1992 Aug;37(4):247–254. doi: 10.1002/jmv.1890370403. [DOI] [PubMed] [Google Scholar]
- Chang C. K., Balachandran N. Identification, characterization, and sequence analysis of a cDNA encoding a phosphoprotein of human herpesvirus 6. J Virol. 1991 Jun;65(6):2884–2894. doi: 10.1128/jvi.65.6.2884-2894.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen G. H., Dietzschold B., Ponce de Leon M., Long D., Golub E., Varrichio A., Pereira L., Eisenberg R. J. Localization and synthesis of an antigenic determinant of herpes simplex virus glycoprotein D that stimulates the production of neutralizing antibody. J Virol. 1984 Jan;49(1):102–108. doi: 10.1128/jvi.49.1.102-108.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhurst S., Chandran B., McIntyre K., Schnabel K., Hall C. B. Phenotypic and genetic polymorphisms among human herpesvirus-6 isolates from North American infants. Virology. 1992 Sep;190(1):490–493. doi: 10.1016/0042-6822(92)91240-u. [DOI] [PubMed] [Google Scholar]
- Eisenberg R. J., Long D., Ponce de Leon M., Matthews J. T., Spear P. G., Gibson M. G., Lasky L. A., Berman P., Golub E., Cohen G. H. Localization of epitopes of herpes simplex virus type 1 glycoprotein D. J Virol. 1985 Feb;53(2):634–644. doi: 10.1128/jvi.53.2.634-644.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnann J. W., Jr, Nelson J. A., Oldstone M. B. Fine mapping of an immunodominant domain in the transmembrane glycoprotein of human immunodeficiency virus. J Virol. 1987 Aug;61(8):2639–2641. doi: 10.1128/jvi.61.8.2639-2641.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett R. F., Gallagher A., Gledhill S., Jones M. D., Teo I., Griffin B. E. Variation in restriction map of MHV-6 genome. Lancet. 1989 Feb 25;1(8635):448–449. doi: 10.1016/s0140-6736(89)90052-4. [DOI] [PubMed] [Google Scholar]
- Jenison S. A., Yu X. P., Valentine J. M., Galloway D. A. Characterization of human antibody-reactive epitopes encoded by human papillomavirus types 16 and 18. J Virol. 1991 Mar;65(3):1208–1218. doi: 10.1128/jvi.65.3.1208-1218.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephs S. F., Ablashi D. V., Salahuddin S. Z., Kramarsky B., Franza B. R., Jr, Pellett P., Buchbinder A., Memon S., Wong-Staal F., Gallo R. C. Molecular studies of HHV-6. J Virol Methods. 1988 Sep;21(1-4):179–190. doi: 10.1016/0166-0934(88)90064-x. [DOI] [PubMed] [Google Scholar]
- Kikuta H., Lu H., Matsumoto S., Josephs S. F., Gallo R. C. Polymorphism of human herpesvirus 6 DNA from five Japanese patients with exanthem subitum. J Infect Dis. 1989 Sep;160(3):550–551. doi: 10.1093/infdis/160.3.550. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Levy J. A., Ferro F., Lennette E. T., Oshiro L., Poulin L. Characterization of a new strain of HHV-6 (HHV-6SF) recovered from the saliva of an HIV-infected individual. Virology. 1990 Sep;178(1):113–121. doi: 10.1016/0042-6822(90)90384-4. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Lopez C., Pellett P., Stewart J., Goldsmith C., Sanderlin K., Black J., Warfield D., Feorino P. Characteristics of human herpesvirus-6. J Infect Dis. 1988 Jun;157(6):1271–1273. doi: 10.1093/infdis/157.6.1271. [DOI] [PubMed] [Google Scholar]
- Marlin S. D., Holland T. C., Levine M., Glorioso J. C. Epitopes of herpes simplex virus type 1 glycoprotein gC are clustered in two distinct antigenic sites. J Virol. 1985 Jan;53(1):128–136. doi: 10.1128/jvi.53.1.128-136.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. E., Thomson B. J., Honess R. W., Craxton M. A., Gompels U. A., Liu M. Y., Littler E., Arrand J. R., Teo I., Jones M. D. The genome of human herpesvirus 6: maps of unit-length and concatemeric genomes for nine restriction endonucleases. J Gen Virol. 1991 Jan;72(Pt 1):157–168. doi: 10.1099/0022-1317-72-1-157. [DOI] [PubMed] [Google Scholar]
- Neipel F., Ellinger K., Fleckenstein B. The unique region of the human herpesvirus 6 genome is essentially collinear with the UL segment of human cytomegalovirus. J Gen Virol. 1991 Sep;72(Pt 9):2293–2297. doi: 10.1099/0022-1317-72-9-2293. [DOI] [PubMed] [Google Scholar]
- Pellett P. E., Black J. B., Yamamoto M. Human herpesvirus 6: the virus and the search for its role as a human pathogen. Adv Virus Res. 1992;41:1–52. doi: 10.1016/s0065-3527(08)60034-2. [DOI] [PubMed] [Google Scholar]
- Salahuddin S. Z., Ablashi D. V., Markham P. D., Josephs S. F., Sturzenegger S., Kaplan M., Halligan G., Biberfeld P., Wong-Staal F., Kramarsky B. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science. 1986 Oct 31;234(4776):596–601. doi: 10.1126/science.2876520. [DOI] [PubMed] [Google Scholar]
- Schirmer E. C., Wyatt L. S., Yamanishi K., Rodriguez W. J., Frenkel N. Differentiation between two distinct classes of viruses now classified as human herpesvirus 6. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5922–5926. doi: 10.1073/pnas.88.13.5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga S., Yoshikawa T., Asano Y., Yazaki T., Hirata S. Human herpesvirus-6 infection (exanthem subitum) without rash. Pediatrics. 1989 Jun;83(6):1003–1006. [PubMed] [Google Scholar]
- Windheuser M. G., Tegtmeier G. E., Wood C. Use of TrpE/Gag fusion proteins to characterize immunoreactive domains on the human immunodeficiency virus type 1 core protein. J Virol. 1989 Sep;63(9):4064–4068. doi: 10.1128/jvi.63.9.4064-4068.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt L. S., Balachandran N., Frenkel N. Variations in the replication and antigenic properties of human herpesvirus 6 strains. J Infect Dis. 1990 Oct;162(4):852–857. doi: 10.1093/infdis/162.4.852. [DOI] [PubMed] [Google Scholar]
- Yamanishi K., Okuno T., Shiraki K., Takahashi M., Kondo T., Asano Y., Kurata T. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet. 1988 May 14;1(8594):1065–1067. doi: 10.1016/s0140-6736(88)91893-4. [DOI] [PubMed] [Google Scholar]