Abstract
Protein folding can be described in terms of the development of specific contacts between residues as a highly disordered polypeptide chain converts into the native state. Here we describe an NMR based strategy designed to detect such contacts by observation of nuclear Overhauser effects (NOEs). Experiments with α-lactalbumin reveal the existence of extensive NOEs between aromatic and aliphatic protons in the archetypal molten globule formed by this protein at low pH. Analysis of their time development provides direct evidence for near-native compactness of this state. Through a rapid refolding procedure the NOE intensity can be transferred efficiently into the resolved and assigned spectrum of the native state. This demonstrates the viability of using this approach to map out time-averaged interactions between residues in a partially folded protein.
One of the most intriguing aspects of protein folding is that the polypeptide chain must develop its structure in a manner which enables it to avoid searching an impossibly large region of conformational space (1, 2). Although the folding of a number of small proteins has been found to be highly cooperative, that of a number of larger ones has been found to involve the initial formation of partially structured states which require a significantly longer time to form the fully native protein (3, 4). This has been taken to indicate that rearrangements within such states may be necessary to allow the protein to achieve a sufficiently native-like structure to progress in folding; reorganization of misfolded species may be an important aspect of this process (5, 6). Such ideas from experimental studies have been supported by simulations of protein folding, particularly using lattice models, which focus on the nature of the contacts formed between residues at different stages of the folding reaction (7–10).
The existence of relatively long-lived species in experimental studies provides the opportunity to develop techniques by which to study their structures (11, 12). A combination of stopped-flow spectroscopic techniques and quenched-flow hydrogen exchange experiments has provided a wealth of information. Recently NMR spectroscopy has also been used in real time to follow aspects of the folding process including the direct observation of kinetic folding and unfolding intermediates (13–16). One result from investigations of an increasing number of proteins by these methods has been support for the concept of the “molten globule” state, a form of a protein which is compact and has extensive secondary structure but which lacks some or all of the persistent tertiary interactions typical of native globular proteins, as a common intermediate in the folding process (17). As well as providing details of kinetic intermediates, these techniques have also provided the key information that stable partially folded species, formed under mildly denaturing conditions and amenable to detailed study, can be good analogues of the transient species (17–19). α-Lactalbumin which forms a well defined molten globule state (the A-state) at low pH is one of the proteins for which this has been shown to be the case (15, 16, 20).
The importance of detecting contacts between residues to define the structural properties of a partially folded state has prompted us to explore methods whereby this information can be obtained. In native proteins nuclear Overhauser effects (NOEs) (21, 22) detected in NMR experiments provide information about distances between individual protons, thus allowing solution structures to be determined. In disordered structures these effects will be averaged over the different interconverting conformations, but will reflect both the number and distribution of residue contacts. Measurement of these averaged NOEs should, therefore, provide crucial information needed to define the nature of a given conformational ensemble. In this paper we describe a strategy to make such measurements, and illustrate it with experiments on the low pH molten globule state of bovine α-lactalbumin (BLA).
MATERIALS AND METHODS
BLA.
BLA was obtained from Sigma (type III) and was used without further purification. NMR samples were prepared by dissolving 1.4 mM BLA in D2O, and adjusting the pH to 2.0 to obtain the A-state, and to 7.0 for native state spectra.
NOE Measurements.
NOE spectra were acquired by saturation of the aromatic resonances (7.0–8.0 ppm) using high-power continuous wave irradiation, and difference spectra were obtained by subtracting reference spectra acquired after saturation at a frequency 2,000 Hz down-field of the aromatic region. NMR spectra were recorded at 20°C using a home-built NMR spectrometer operating at 600.2 MHz, and processed using felix (Biosym Technologies, San Diego). The residual water resonance was saturated by weak on-resonance irradiation during the 1-s relaxation delay. The spectral width was 9,615 Hz and a 90° shifted squared sinebell window function was applied before Fourier transformation.
NOE Buildup and Decay Curves.
Aliphatic NOE intensities were obtained by integrating NOE difference spectra over the aliphatic region, up-field of 3.5 ppm. The NOE build-up was measured by varying the saturation time between 0 and 1,200 ms, while its decay was measured using a 1,200-ms saturation time followed by a delay of between 0 and 800 ms. Build-up curves were fitted by a two-spin model for which the NOE intensity (21, 22), η, is given by η = σ/ρ [1 − exp(−ρt)], where σ is the cross relaxation rate and ρ is the spin-lattice relaxation rate. Decay curves were (arbitrarily) fitted to a single exponential.
Refolding Experiments.
Rapid refolding of BLA within an NMR tube was accomplished by mixing 55 μl of 330 mM Tris (pH 8.9) and 400 mM Ca2+ into 450 μl of 20 mg/ml BLA in D2O at pH 2.0 using a pneumatic injection device (15). NOE spectra were obtained by saturating the aromatic region in the A-state for 1 s, followed by rapid refolding; the NOE was detected 800 ms later, by which time mixing and refolding are complete [the half-time for the refolding process is 30 ms (23)]. Difference spectra were obtained by subtracting reference spectra as described above. The experiments were repeated 12 times to improve the signal-to-noise ratios.
RESULTS AND DISCUSSION
Generation of NOEs in the A-State.
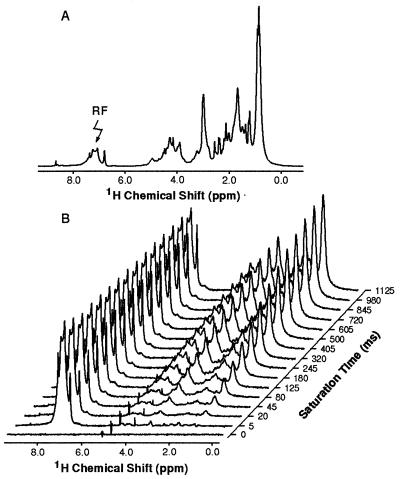
Fig. 1a shows the one-dimensional NMR spectrum of α-lactalbumin in its A-state formed at pH 2.0. The spectrum shows the broad lines and limited spectral dispersion familiar from studies of partially folded proteins, and attributable primarily to slow rearrangements of the structure (17, 24). Such one-dimensional experiments, however, provide an opportunity to probe the overall pattern of NOEs arising from particular groups of protons in the form of intensity changes resulting from saturation of their resonances. It is apparent from the spectrum in Fig. 1a that the aromatic proton resonances are well separated from others in the spectrum. Moreover, these residues are largely located in the core of the native protein and appear to be key elements in the formation of the compact state (24, 25). Fig. 1b shows the effect of saturating the envelope of aromatic resonances on the intensities of other peaks in the spectrum of the A-state. This reveals that the aromatic region of the spectrum is completely saturated even with the shortest pulse lengths used (5 ms). Moreover, it shows that transfer of saturation takes place to resonances in the aliphatic region of the spectrum, the extent of transfer increasing as the saturation time increases. This transfer must arise as a consequence of cross-relaxation between the aromatic and aliphatic protons—i.e., as a result of NOEs.
Figure 1.
One-dimensional 1H-NMR spectra of BLA in D2O at pH 2.0. (a) The full one-dimensional spectrum of BLA. (b) Difference spectra obtained from BLA following saturation of the aromatic resonances (7.0–8.0 ppm) by high power continuous wave irradiation at the position labeled by an arrow in a.
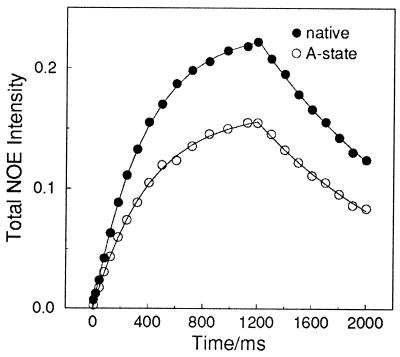
The initial rate of build-up of the NOE intensity in the aliphatic region is determined by the rates of cross-relaxation with the aromatic protons (21, 22). As these rates are distance dependent, their similarity in the native and A-states (Fig. 2) indicates that the A-state is compact and that many close contacts exist between aromatic rings and the side-chains of other residues in the A-state. Substantially smaller effects (Fig. 3) are seen when the experiment is repeated for the denatured state of the protein in deuterated 6 M guanidine hydrochloride. The initial rate of NOE development is less than 20% of that observed for the A-state, consistent with the highly unfolded nature of the protein under these conditions. The residual NOE effects can be attributed to cross-relaxation between protons of residues close to each other in the protein sequence and to the existence of residual local structure known to be present, particularly in the vicinity of aromatic residues, even in otherwise highly unfolded states (26).
Figure 2.
Build-up and decay curves for NOE intensities in different states of BLA at 20°C. The apparent cross-relaxation rates (σ) measured were 0.62 ± 0.01 s−1 for the native state and 0.41 ± 0.01 s−1 for the A-state; corresponding spin-lattice relaxation rates (ρ) were 2.67 ± 0.05 s−1 and 2.49 ± 0.08 s−1, respectively. The inversion-recovery spin-lattice relaxation rate for the native state was measured as 2.20 ± 0.08 s−1; this rate, however, is averaged over all aliphatic protons, while the build-up rate is only averaged over those which experience substantial NOEs.
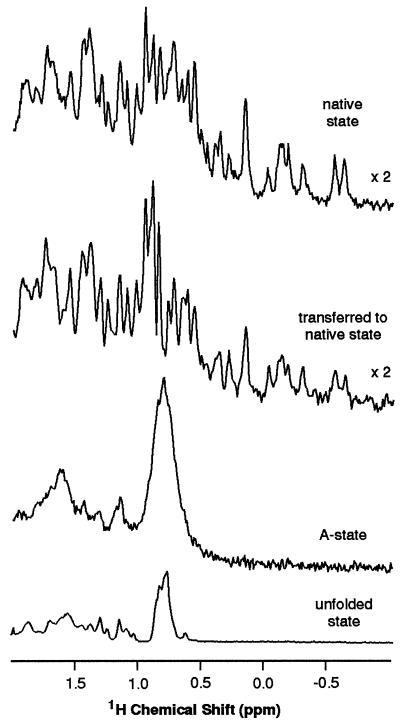
Figure 3.
Up-field aliphatic spectral region of NOE difference spectra of BLA in D2O at 20°C; spectra are shown for the native, refolded, A-, and unfolded (6 M guanidine hydrochloride) states. Substantial NOEs are observed for the native and A-states, but only a small NOE is observed for the unfolded state. The spectrum for the refolded state shows the effect of generating NOEs in the A-state and transferring them to the native state for detection.
Detailed analysis of these build-up rates is complicated, as the cross-relaxation rate depends on both the time-averaged values of proton–proton distances and the details of the motional processes which give rise to relaxation (21, 22). We assume a simple model in which an increase in volume of the A-state increases the average proton–proton distance and increases the rotational correlation time of the protein. The homonuclear cross-relaxation rate depends on the inverse sixth power of the internuclear distance and the motional spectral density at zero frequency; for a sphere undergoing isotropic tumbling this spectral density increases with the cube of the radius. Combining these two effects suggests that the cross-relaxation rate should vary with the inverse cube of the molecular radius. The effects of averaging, of local rapid motions, and of intraresidue cross-relaxation have been ignored. Using this model, the ratio between the cross-relaxation rates of the A-state and the native state (0.66 ± 0.02) indicates that the effective radius of the protein increases by 15 ± 2% in the A-state, a value highly consistent with the 10–20% reported previously based on viscosity measurements (20, 27).
Transfer of NOEs to the Native State.
Although the results discussed above indicate that the time-averaged number of contacts (at least between aliphatic and aromatic protons) within the ensemble of structures contributing to the A-state is comparable to that of the native state, the very low resolution of the A-state spectrum means that the experiment does not provide information about the specific nature of these contacts. To overcome this limitation we have devised the following strategy. After the generation of NOEs in the A-state, the protein is rapidly mixed with buffer to produce an environment in which it can fold to the native state; when this happens, the magnetization resulting from the saturation in the A-state is transferred to the native state. The principle is analogous to that of hydrogen exchange experiments (28, 29) where isotope, rather than nuclear spin, labeling is transferred from an intermediate to the native state for detection. As long as refolding can be accomplished before the NOE intensity has decayed away, the NOE generated in the A-state should then be detectable in the high resolution spectrum of the native state. The NOE decay rates shown in Fig. 2 suggest that our refolding time of 800 ms is short enough to allow such transferred NOEs to be detected. This is confirmed by the experimental results for BLA shown in Fig. 3. After saturation of the aromatic resonances of the A-state followed by refolding, a substantial NOE intensity is indeed seen in the aliphatic region of the native spectrum indicative of NOEs generated in the A-state. Integration of the spectra up-field of 3.5 ppm indicates that the NOE intensity recorded in the native spectrum is more than 65% of that in the A-state prior to the transfer procedure.
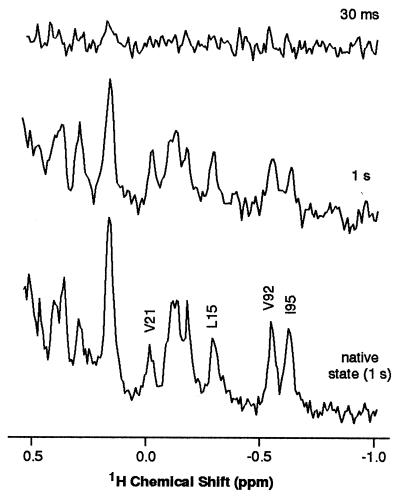
During the refolding period any given protein molecule spends a fraction of its time in the native state, and so the NOE intensity will partly reflect relaxation behavior in the native state. To test the consequences of this, experiments were carried out with saturation times sufficiently short that the build up of NOEs in the A-state prior to the mixing procedure will be negligible. Substantially reduced intensity changes resulted; with a 30-ms pulse less than 10% of the NOE in the 1-s saturation experiment was observed (Fig. 4). This indicates that the observed NOE intensities in the experiments reported here predominantly reflect the NOEs generated in the A-state. Improvements in the mixing procedure allowing the use of shorter refolding times would further reduce any contributions from the native state. Even with the present mixing device, however, the NOE intensities measured in the high resolution spectrum of the native protein can be seen to monitor within experimental error the time-averaged contacts between aromatic and aliphatic protons in the partially folded state. These results, therefore, show the potential of this approach to determine interactions between residues even in a state where the NMR spectrum is very poorly resolved.
Figure 4.
Expansions of the high-field regions of NOE difference spectra of BLA in D2O at 20°C; spectra are shown for the native state and for the refolded state after saturation of the A-state aromatic protons for 30 ms and 1 s. Assigned residues (R. Wijesinha & C. Redfield, personal communication) are indicated by their one letter codes (V, valine; L, leucine; I, isoleucine).
In the experiments described in this paper the broad envelope of aromatic resonances was saturated to generate the NOEs. This means that the specific aromatic protons giving rise to the NOE on a given aliphatic proton are not known; the present experiments do not therefore allow information about individual contacts to be determined. They do, however, allow some preliminary conclusions to be drawn about the nature of the contacts within the A-state by comparing the intensities of these NOEs with those measured for the native state using the same saturation conditions. Such a comparison is shown in Figs. 3 and 4. It is apparent that there is a general similarity between the intensity distribution in the NOEs generated in the two states. Some qualitative differences do, however, stand out. Most notably, the NOEs detected in the resonances in the high field region of the spectrum, up-field of 0.5 ppm, are significantly smaller in the A-state than the native state, while NOEs detected in resonances close to 1.0 ppm are relatively more intense.
These resonances in the high-field region of the spectrum are all of methyl groups close to aromatic residues in the native structure; the ring current shifts arising from these proximities are the origin of their large shift perturbations from random coil values. As significant NOEs are only likely be observed under saturation conditions such as those used here between protons that approach closer than about 5 Å (30), the intensities of the native state NOEs on these resonances are large compared with those on the resonances of the majority of the aliphatic protons. The generally reduced magnitude of the NOEs in the A-state for these methyl groups suggests, therefore, that on average their proximities to aromatic residues are less than in the native state. Such a result is consistent with the loss of specific close packing in the A-state indicated by the reduced chemical shift dispersion in the NMR spectrum (24) and the almost complete loss of near-UV CD signal (20, 27) characteristic of ordered aromatic residues. By contrast, the increased intensity in the A-state NOE spectrum close to 1.0 ppm, the region of the spectrum where methyl groups distant from aromatic rings in the native state are expected to resonate, suggests the existence of nonnative contacts between methyl groups and aromatic residues.
Although NOE intensities in the high-field region of the spectrum are generally reduced, the magnitude of this reduction varies for different resonances. One case where the NOE appears almost identically in the native and A-states is Val-21 (see Fig. 4). In the native structure this residue is located at the N terminus of one of the four α-helices, helix B, and the aromatic residue closest to it is Trp-26, itself located in the B-helix [we used a model of BLA as described in reference (15)]. The B-helix has been shown from hydrogen exchange and proline mutagenesis experiments to be stable in the A-state of α-lactalbumin (25, 31). Moreover, the 28–111 disulfide bond has been found to form highly efficiently during oxidative folding, suggesting a strong bias toward the native fold in this region (32). The native-like intensity of the observed NOE would therefore be consistent with the maintenance of native-like proximities in this region. By contrast, the NOE experienced by the up-field methyl group of Ile-95 is reduced by a factor of about 2 in the A-state relative to the native state. This residue is a part of helix C in the native structure, which is also persistent in the A-state (24, 25, 33). The closest aromatic residue to Ile-95 in the native structure is, however, Tyr-103, which is located close to the D-helix. Previous experiments have indicated that the side chain of this residue is in a nonnative conformation in the A-state (33); such a change would be consistent with the change in NOE intensity observed here. However, further studies will be necessary to confirm these suggestions.
CONCLUSIONS
The experiments described in this paper indicate that an overall strategy to probe the tertiary structural properties of partially folded states by detection of NOEs is viable. A particularly important finding is that NOEs between residues in the A-state are similar in overall intensity to those in the native state. Earlier attempts to detect such NOEs in the A-state directly using two-dimensional NMR experiments were, however, largely unsuccessful (33). The present results suggest that this is due to the low intensity of cross-peaks arising from broad resonances, particularly when their specificity is reduced in a disordered state, rather than from a lack of close interactions between residues. The experiments described in this paper, by which the NOEs are generated in a partially folded state but observed in the native state, provide a means of overcoming this problem. Although the approach has not yet permitted contacts between individual residues to be defined, such information could in principle be achieved by increasing the selectivity of the experiment, for example through isotope labeling or site-directed mutagenesis. Under such circumstances the time-averaged proximities of specific groups within a partially folded protein could in principle be determined. Moreover, the approach could be coupled with additional mixing experiments to probe the distribution of contacts within kinetically generated species present at different stages of the folding process. This would then provide an opportunity to provide experimental information to begin to map out the energy landscapes (2) which describe the manner in which protein molecules fold.
Acknowledgments
We thank Steve L. Winder and Peter J. Hore for help in developing the rapid mixing device. We thank Christina Redfield for helpful suggestions. J.B., V.F., and N.A.J.v.N. were supported by the Human Capital and Mobility Program of the European Community. J.A.J. thanks Merton College Oxford for a Junior Research Fellowship. This work is a contribution from the Oxford Centre for Molecular Sciences which is supported by the Engineering and Physical Sciences Research Council, the Biotechnology and Biological Sciences Research Council, and the Medical Research Council. The research of C.M.D. is supported in part by an International Research Scholars award from the Howard Hughes Medical Institute.
ABBREVIATIONS
- NOE
nuclear Overhauser effect
- BLA
bovine α-lactalbumin
References
- 1.Levinthal C. J Chim Phys. 1968;65:44–45. [Google Scholar]
- 2.Wolynes P G, Onuchic J N, Thirumalai D. Science. 1995;268:960–961. doi: 10.1126/science.268.5213.960. [DOI] [PubMed] [Google Scholar]
- 3.Fersht A R. Proc Natl Acad Sci USA. 1995;92:10869–10873. doi: 10.1073/pnas.92.24.10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miranker A D, Dobson C M. Curr Opin Struct Biol. 1996;6:31–42. doi: 10.1016/s0959-440x(96)80092-3. [DOI] [PubMed] [Google Scholar]
- 5.Radford S E, Dobson C M, Evans P A. Nature (London) 1992;358:302–307. doi: 10.1038/358302a0. [DOI] [PubMed] [Google Scholar]
- 6.Sosnick T R, Mayne L, Miller R, Englander S W. Nat Struct Biol. 1994;1:149–156. doi: 10.1038/nsb0394-149. [DOI] [PubMed] [Google Scholar]
- 7.Chan H S, Bromberg S, Dill K A. Philos Trans R Soc London. 1995;348:61–70. doi: 10.1098/rstb.1995.0046. [DOI] [PubMed] [Google Scholar]
- 8.Onuchic J N, Wolynes P G, Luthey-Schulten Z, Socci N D. Proc Natl Acad Sci USA. 1995;92:3626–3630. doi: 10.1073/pnas.92.8.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karplus M, Sali A. Curr Opin Struct Biol. 1995;4:58–73. doi: 10.1016/0959-440x(95)80010-x. [DOI] [PubMed] [Google Scholar]
- 10.Thirumalai D, Woodson S A. Acc Chem Res. 1996;29:433–439. [Google Scholar]
- 11.Evans P A, Radford S E. Curr Opin Struct Biol. 1994;4:100–106. [Google Scholar]
- 12.Plaxco K W, Dobson C M. Curr Opin Struct Biol. 1996;6:630–636. doi: 10.1016/s0959-440x(96)80029-7. [DOI] [PubMed] [Google Scholar]
- 13.Hoeltzli S D, Frieden C. Proc Natl Acad Sci USA. 1995;92:9318–9322. doi: 10.1073/pnas.92.20.9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiefhaber T, Labhardt A M, Baldwin R L. Nature (London) 1995;375:513–515. doi: 10.1038/375513a0. [DOI] [PubMed] [Google Scholar]
- 15.Balbach J, Forge V, van Nuland N A J, Winder S L, Hore P J, Dobson C M. Nat Struct Biol. 1995;2:865–870. doi: 10.1038/nsb1095-865. [DOI] [PubMed] [Google Scholar]
- 16.Balbach J, Forge V, Lau W S, van Nuland N A J, Brew K, Dobson C M. Science. 1996;274:1161–1163. doi: 10.1126/science.274.5290.1161. [DOI] [PubMed] [Google Scholar]
- 17.Ptitsyn O B. Curr Opin Struct Biol. 1995;5:74–78. doi: 10.1016/0959-440x(95)80011-o. [DOI] [PubMed] [Google Scholar]
- 18.Jennings P A, Wright P E. Science. 1993;262:892–896. doi: 10.1126/science.8235610. [DOI] [PubMed] [Google Scholar]
- 19.Fink A L. Annu Rev Biophys Biomol Struct. 1995;24:495–522. doi: 10.1146/annurev.bb.24.060195.002431. [DOI] [PubMed] [Google Scholar]
- 20.Kuwajima K. FASEB J. 1996;10:102–109. doi: 10.1096/fasebj.10.1.8566530. [DOI] [PubMed] [Google Scholar]
- 21.Noggle J H, Schirmer R F. The Nuclear Overhauser Effect: Chemical Applications. New York: Academic; 1971. [Google Scholar]
- 22.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 23.Ikeguchi M, Kuwajima K, Mitani M, Sugai S. Biochemistry. 1986;25:6965–6972. doi: 10.1021/bi00370a034. [DOI] [PubMed] [Google Scholar]
- 24.Baum J, Dobson C M, Evans P A, Hanley C. Biochemistry. 1989;28:7–13. doi: 10.1021/bi00427a002. [DOI] [PubMed] [Google Scholar]
- 25.Schulman B A, Kim P S. Nat Struct Biol. 1996;3:602–607. doi: 10.1038/nsb0896-682. [DOI] [PubMed] [Google Scholar]
- 26.Broadhurst R W, Dobson C M, Hore P J, Radford S E, Rees M L. Biochemistry. 1991;30:405–412. doi: 10.1021/bi00216a015. [DOI] [PubMed] [Google Scholar]
- 27.Dolgikh D A, Gilmanshin R I, Brazhnikov E V, Bychkova V E, Semisotnov G V, Venyaminov S Y, Ptitsyn O B. FEBS Lett. 1981;136:311–315. doi: 10.1016/0014-5793(81)80642-4. [DOI] [PubMed] [Google Scholar]
- 28.Englander S W, Mayne L. Annu Rev Biophys Biomol Struct. 1992;21:243–265. doi: 10.1146/annurev.bb.21.060192.001331. [DOI] [PubMed] [Google Scholar]
- 29.Baldwin R L. Curr Opin Struct Biol. 1993;3:84–91. [Google Scholar]
- 30.Poulsen F M, Hoch J C, Dobson C M. Biochemistry. 1980;19:2597–2607. doi: 10.1021/bi00553a011. [DOI] [PubMed] [Google Scholar]
- 31.Schulman B A, Kim P S. Nat Struct Biol. 1996;3:682–687. doi: 10.1038/nsb0896-682. [DOI] [PubMed] [Google Scholar]
- 32.Wu L C, Peng Z Y, Kim P S. Nat Struct Biol. 1995;2:281–286. doi: 10.1038/nsb0495-281. [DOI] [PubMed] [Google Scholar]
- 33.Alexandrescu A T, Evans P A, Pitkeathly M, Baum J, Dobson C M. Biochemistry. 1993;32:1707–1718. doi: 10.1021/bi00058a003. [DOI] [PubMed] [Google Scholar]