Abstract
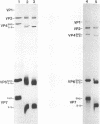
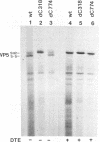
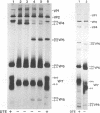
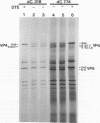
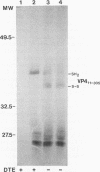
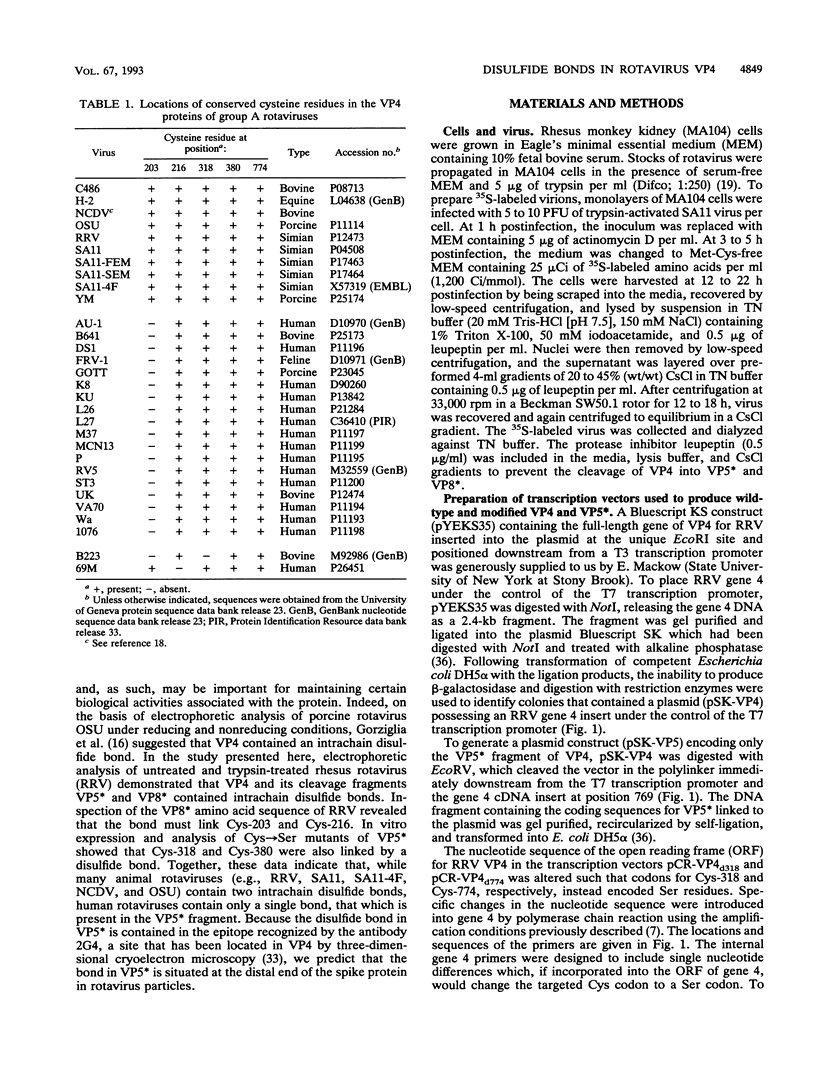
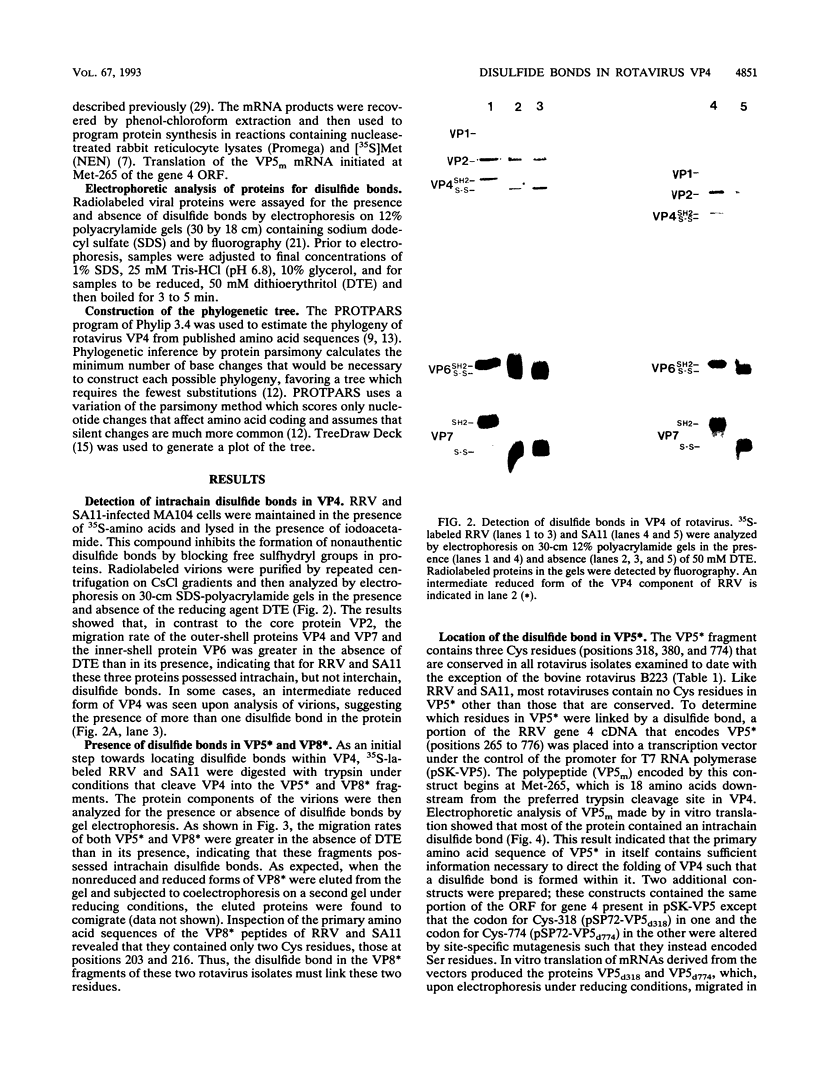
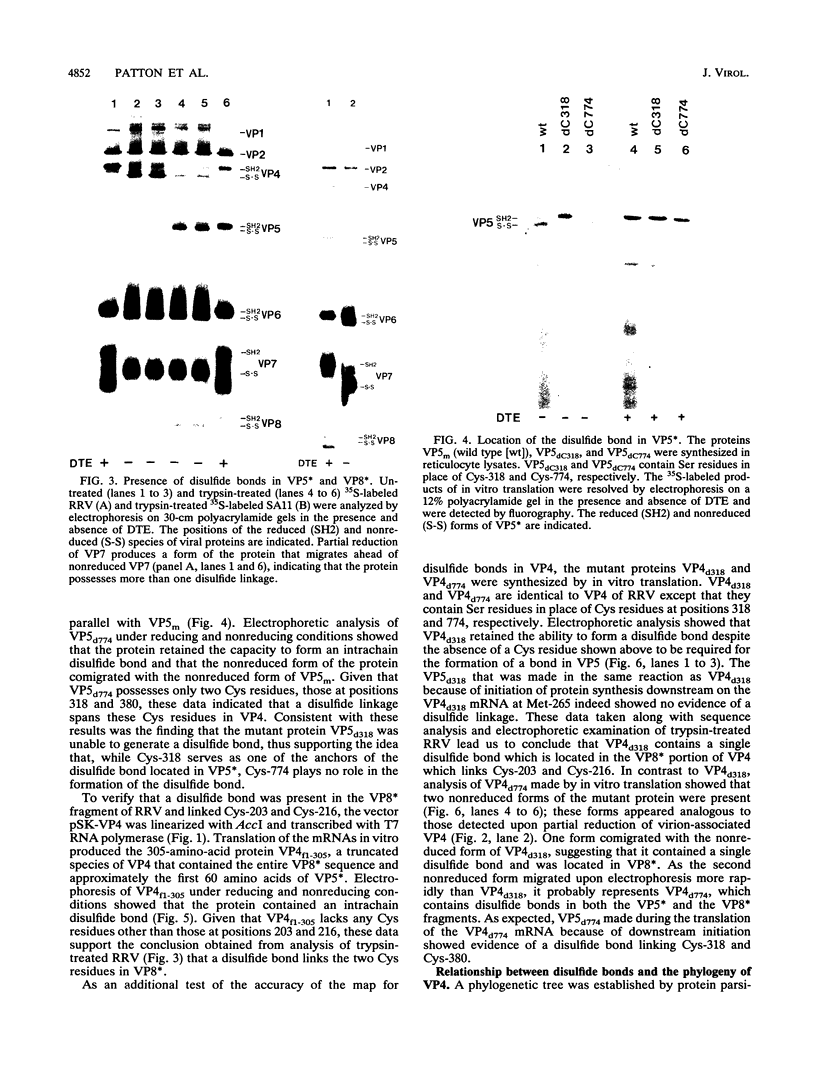
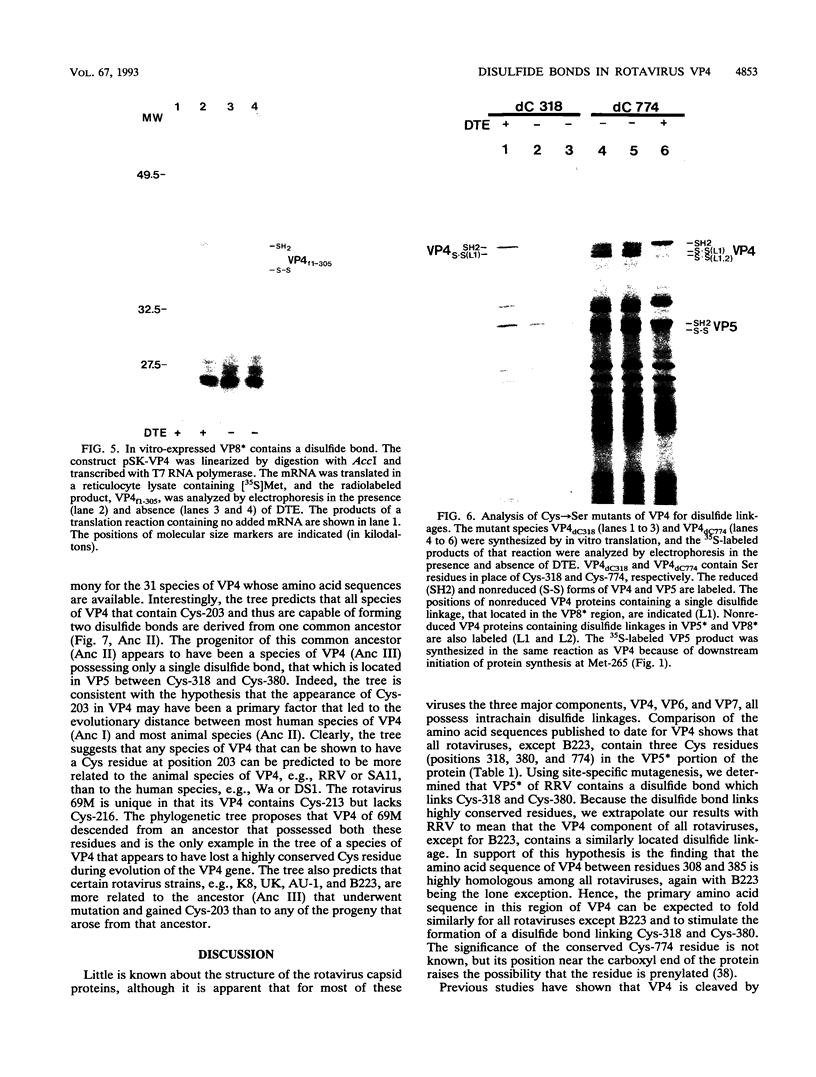
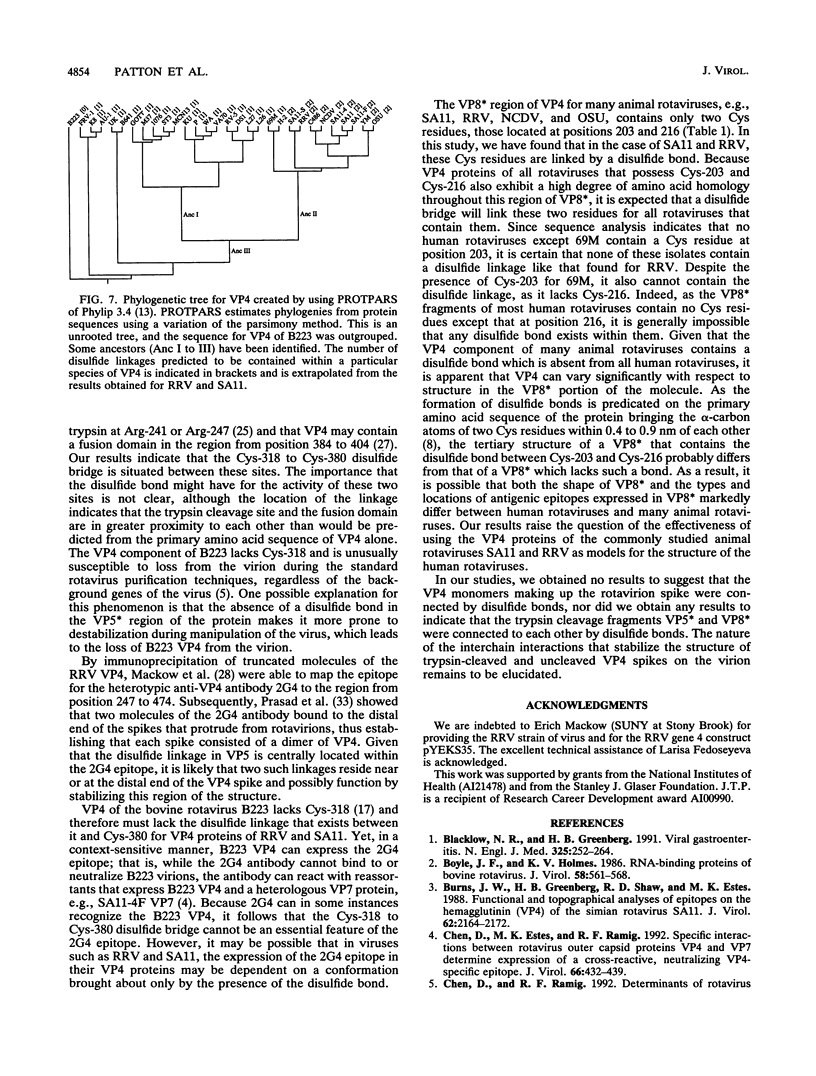
Because the rotavirus spike protein VP4 contains conserved Cys residues at positions 216, 318, 380, and 774 and, for many animal rotaviruses, also at position 203, we sought to determine whether disulfide bonds were structural elements of VP4. Electrophoretic analysis of untreated and trypsin-treated rhesus rotavirus (RRV) and simain rotavirus SA11 in the presence and absence of the reducing agent dithioerythritol revealed that VP4 and its cleavage fragments VP5* and VP8* possessed intrachain disulfide bonds. Given that the VP8* fragments of RRV and SA11 contain only two Cys residues, those at positions 203 and 216, these data indicated that these two residues were covalently linked. Electrophoretic examination of truncated species of VP4 and VP4 containing Cys-->Ser mutations synthesized in reticulocyte lysates provided additional evidence that Cys-203 and Cys-216 in VP8* of RRV were linked by a disulfide bridge. VP5* expressed in vitro was able to form a disulfide bond analogous to that in the VP5* fragment of trypsin-treated RRV. Analysis of a Cys-774-->Ser mutant of VP5* showed that, while it was able to form a disulfide bond, a Cys-318-->Ser mutant of VP5* was not. These results indicated that the VP4 component of all rotaviruses, except B223, contains a disulfide bond that links Cys-318 and Cys-380 in the VP5* region of the protein. This bond is located between the trypsin cleavage site and the putative fusion domain of VP4. Because human rotaviruses lack Cys-203 and, hence, unlike many animal rotaviruses cannot possess a disulfide bond in VP8*, it is apparent that VP4 is structurally variable in nature, with human rotaviruses generally containing one disulfide linkage and animal rotaviruses generally containing two such linkages. Considered with the results of anti-VP4 antibody mapping studies, the data suggest that the disulfide bond in VP5* exists within the 2G4 epitope and may be located at the distal end of the VP4 spike on rotavirus particles.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blacklow N. R., Greenberg H. B. Viral gastroenteritis. N Engl J Med. 1991 Jul 25;325(4):252–264. doi: 10.1056/NEJM199107253250406. [DOI] [PubMed] [Google Scholar]
- Boyle J. F., Holmes K. V. RNA-binding proteins of bovine rotavirus. J Virol. 1986 May;58(2):561–568. doi: 10.1128/jvi.58.2.561-568.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns J. W., Greenberg H. B., Shaw R. D., Estes M. K. Functional and topographical analyses of epitopes on the hemagglutinin (VP4) of the simian rotavirus SA11. J Virol. 1988 Jun;62(6):2164–2172. doi: 10.1128/jvi.62.6.2164-2172.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D. Y., Estes M. K., Ramig R. F. Specific interactions between rotavirus outer capsid proteins VP4 and VP7 determine expression of a cross-reactive, neutralizing VP4-specific epitope. J Virol. 1992 Jan;66(1):432–439. doi: 10.1128/jvi.66.1.432-439.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D. Y., Ramig R. F. Determinants of rotavirus stability and density during CsCl purification. Virology. 1992 Jan;186(1):228–237. doi: 10.1016/0042-6822(92)90077-3. [DOI] [PubMed] [Google Scholar]
- Christensen M. L. Human viral gastroenteritis. Clin Microbiol Rev. 1989 Jan;2(1):51–89. doi: 10.1128/cmr.2.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapp L. L., Patton J. T. Rotavirus morphogenesis: domains in the major inner capsid protein essential for binding to single-shelled particles and for trimerization. Virology. 1991 Feb;180(2):697–708. doi: 10.1016/0042-6822(91)90083-n. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Disulfide bond formation in proteins. Methods Enzymol. 1984;107:305–329. doi: 10.1016/0076-6879(84)07021-x. [DOI] [PubMed] [Google Scholar]
- Estes M. K., Cohen J. Rotavirus gene structure and function. Microbiol Rev. 1989 Dec;53(4):410–449. doi: 10.1128/mr.53.4.410-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes M. K., Graham D. Y., Mason B. B. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J Virol. 1981 Sep;39(3):879–888. doi: 10.1128/jvi.39.3.879-888.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- Fiore L., Greenberg H. B., Mackow E. R. The VP8 fragment of VP4 is the rhesus rotavirus hemagglutinin. Virology. 1991 Apr;181(2):553–563. doi: 10.1016/0042-6822(91)90888-i. [DOI] [PubMed] [Google Scholar]
- Gorziglia M., Larrea C., Liprandi F., Esparza J. Biochemical evidence for the oligomeric (possibly trimeric) structure of the major inner capsid polypeptide (45K) of rotaviruses. J Gen Virol. 1985 Sep;66(Pt 9):1889–1900. doi: 10.1099/0022-1317-66-9-1889. [DOI] [PubMed] [Google Scholar]
- Hardy M. E., Gorziglia M., Woode G. N. Amino acid sequence analysis of bovine rotavirus B223 reveals a unique outer capsid protein VP4 and confirms a third bovine VP4 type. Virology. 1992 Nov;191(1):291–300. doi: 10.1016/0042-6822(92)90191-q. [DOI] [PubMed] [Google Scholar]
- Hardy M. E., Woode G. N., Xu Z. C., Gorziglia M. Comparative amino acid sequence analysis of VP4 for VP7 serotype 6 bovine rotavirus strains NCDV, B641, and UK. J Virol. 1991 Oct;65(10):5535–5538. doi: 10.1128/jvi.65.10.5535-5538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmberger-Jones M., Patton J. T. Characterization of subviral particles in cells infected with simian rotavirus SA11. Virology. 1986 Dec;155(2):655–665. doi: 10.1016/0042-6822(86)90225-4. [DOI] [PubMed] [Google Scholar]
- Labbé M., Charpilienne A., Crawford S. E., Estes M. K., Cohen J. Expression of rotavirus VP2 produces empty corelike particles. J Virol. 1991 Jun;65(6):2946–2952. doi: 10.1128/jvi.65.6.2946-2952.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larralde G., Li B. G., Kapikian A. Z., Gorziglia M. Serotype-specific epitope(s) present on the VP8 subunit of rotavirus VP4 protein. J Virol. 1991 Jun;65(6):3213–3218. doi: 10.1128/jvi.65.6.3213-3218.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizano M., López S., Arias C. F. The amino-terminal half of rotavirus SA114fM VP4 protein contains a hemagglutination domain and primes for neutralizing antibodies to the virus. J Virol. 1991 Mar;65(3):1383–1391. doi: 10.1128/jvi.65.3.1383-1391.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López S., Arias C. F., Bell J. R., Strauss J. H., Espejo R. T. Primary structure of the cleavage site associated with trypsin enhancement of rotavirus SA11 infectivity. Virology. 1985 Jul 15;144(1):11–19. doi: 10.1016/0042-6822(85)90300-9. [DOI] [PubMed] [Google Scholar]
- López S., López I., Romero P., Méndez E., Soberón X., Arias C. F. Rotavirus YM gene 4: analysis of its deduced amino acid sequence and prediction of the secondary structure of the VP4 protein. J Virol. 1991 Jul;65(7):3738–3745. doi: 10.1128/jvi.65.7.3738-3745.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackow E. R., Barnett J. W., Chan H., Greenberg H. B. The rhesus rotavirus outer capsid protein VP4 functions as a hemagglutinin and is antigenically conserved when expressed by a baculovirus recombinant. J Virol. 1989 Apr;63(4):1661–1668. doi: 10.1128/jvi.63.4.1661-1668.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackow E. R., Shaw R. D., Matsui S. M., Vo P. T., Dang M. N., Greenberg H. B. The rhesus rotavirus gene encoding protein VP3: location of amino acids involved in homologous and heterologous rotavirus neutralization and identification of a putative fusion region. Proc Natl Acad Sci U S A. 1988 Feb;85(3):645–649. doi: 10.1073/pnas.85.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackow E. R., Yamanaka M. Y., Dang M. N., Greenberg H. B. DNA amplification-restricted transcription-translation: rapid analysis of rhesus rotavirus neutralization sites. Proc Natl Acad Sci U S A. 1990 Jan;87(2):518–522. doi: 10.1073/pnas.87.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offit P. A., Shaw R. D., Greenberg H. B. Passive protection against rotavirus-induced diarrhea by monoclonal antibodies to surface proteins vp3 and vp7. J Virol. 1986 May;58(2):700–703. doi: 10.1128/jvi.58.2.700-703.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton J. T., Salter-Cid L., Kalbach A., Mansell E. A., Kattoura M. Nucleotide and amino acid sequence analysis of the rotavirus nonstructural RNA-binding protein NS35. Virology. 1993 Feb;192(2):438–446. doi: 10.1006/viro.1993.1059. [DOI] [PubMed] [Google Scholar]
- Pizarro J. L., Sandino A. M., Pizarro J. M., Fernández J., Spencer E. Characterization of rotavirus guanylyltransferase activity associated with polypeptide VP3. J Gen Virol. 1991 Feb;72(Pt 2):325–332. doi: 10.1099/0022-1317-72-2-325. [DOI] [PubMed] [Google Scholar]
- Prasad B. V., Burns J. W., Marietta E., Estes M. K., Chiu W. Localization of VP4 neutralization sites in rotavirus by three-dimensional cryo-electron microscopy. Nature. 1990 Feb 1;343(6257):476–479. doi: 10.1038/343476a0. [DOI] [PubMed] [Google Scholar]
- Prasad B. V., Wang G. J., Clerx J. P., Chiu W. Three-dimensional structure of rotavirus. J Mol Biol. 1988 Jan 20;199(2):269–275. doi: 10.1016/0022-2836(88)90313-0. [DOI] [PubMed] [Google Scholar]
- Sabara M., Ready K. F., Frenchick P. J., Babiuk L. A. Biochemical evidence for the oligomeric arrangement of bovine rotavirus nucleocapsid protein and its possible significance in the immunogenicity of this protein. J Gen Virol. 1987 Jan;68(Pt 1):123–133. doi: 10.1099/0022-1317-68-1-123. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinensky M., Lutz R. J. The prenylation of proteins. Bioessays. 1992 Jan;14(1):25–31. doi: 10.1002/bies.950140106. [DOI] [PubMed] [Google Scholar]
- Valenzuela S., Pizarro J., Sandino A. M., Vásquez M., Fernández J., Hernández O., Patton J., Spencer E. Photoaffinity labeling of rotavirus VP1 with 8-azido-ATP: identification of the viral RNA polymerase. J Virol. 1991 Jul;65(7):3964–3967. doi: 10.1128/jvi.65.7.3964-3967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager M., Dryden K. A., Olson N. H., Greenberg H. B., Baker T. S. Three-dimensional structure of rhesus rotavirus by cryoelectron microscopy and image reconstruction. J Cell Biol. 1990 Jun;110(6):2133–2144. doi: 10.1083/jcb.110.6.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]