Abstract
The phosphotyrosine-binding (PTB) domain is a recently identified protein module that has been characterized as binding to phosphopeptides containing an NPXpY motif (X = any amino acid). We describe here a novel peptide sequence recognized by the PTB domain from Drosophila Numb (dNumb), a protein involved in cell fate determination and asymmetric cell division during the development of the Drosophila nervous system. Using a Tyr-oriented peptide library to screen for ligands, the dNumb PTB domain was found to bind selectively to peptides containing a YIGPYφ motif (φ represents a hydrophobic residue). A synthetic peptide containing this sequence bound specifically to the isolated dNumb PTB domain in solution with a dissociation constant (Kd) of 5.78 ± 0.74 μM. Interestingly, the affinity of this peptide for the dNumb PTB domain was increased (Kd = 1.41 ± 0.10 μM) when the second tyrosine in the sequence was phosphorylated. Amino acid substitution studies of the phosphopeptide demonstrated that a core motif of sequence GP(p)Y is required for high-affinity binding to the dNumb PTB domain. Nuclear magnetic resonance experiments performed on isotopically labeled protein complexed with either Tyr- or pTyr-containing peptides suggest that the same set of amino acids in the dNumb PTB domain is involved in binding both phosphorylated and nonphosphorylated forms of the peptide. The in vitro selectivity of the dNumb PTB domain is therefore markedly different from those of the Shc and IRS-1 PTB domains, in that it interacts preferentially with a GP(p)Y motif, rather than NPXpY, and does not absolutely require ligand phosphorylation for binding. Our results suggest that the PTB domain is a versatile protein module, capable of exhibiting varied binding specificities.
Tyrosine phosphorylation and dephosphorylation are important means of regulating cellular responses to external cues (1). Activation of a receptor tyrosine kinase by binding of an extracelluar ligand generally results in phosphorylation of specific tyrosine residues within the receptor’s cytoplasmic domain, which, in turn, serve as binding sites for downstream signaling molecules containing Src homology 2 (SH2) domains (2, 3). Recently, a distinct protein module capable of binding to phosphotyrosine (pTyr) sites was found in several proteins, including the adaptor protein Shc, and the insulin receptor substrates IRS-1 and IRS-2 (4–9). This module, termed the phosphotyrosine-binding (PTB; refs. 4, 6, and 7) or phosphotyrosine-interaction (PI; refs. 5 and 10) domain differs from the SH2 domain in that it recognizes phosphotyrosine in the sequence context NPXpY, in contrast to SH2 domains, which recognize residues C-terminal to pTyr (2, 11). Different PTB domains exhibit distinct selectivity for residues at positions −5 to −8 N-terminal to the pTyr (6, 7, 12, 13). For instance, the Shc PTB domain requires a hydrophobic residue at pTyr-5 for high-affinity binding, whereas the IRS-1 PTB domain favors hydrophobic residues at the pTyr-6 and pTyr-8 positions (6, 7, 13). The structural basis for such variations in specificity was recently elucidated by nuclear magnetic resonance (NMR) studies on the Shc PTB domain in complex with a TrkA phosphopeptide (14), and by x-ray and NMR analysis of the IRS-1 PTB domain complexed with phosphopeptides derived from the insulin receptor (IR) and the IL-4 receptor, respectively (15, 16).
Sequence alignment has revealed that several proteins other than Shc and IRS-1 also contain potential PTB domains (10). These include the neuronal proteins Fe65 and X11, and a membrane-associated protein, Numb. Although it is now well documented that the PTB domains of Shc and IRS-1 bind selectively to the NPXpY sequence, it seems probable that these other PTB domains may have distinct binding specificity. It has been reported that the PTB domain of X11 and the C-terminal PTB domain of FE65 can interact with the intracellular domain of the β-amyloid precursor protein (β-APP) (17, 18). The PTB-domain-binding site on the β-amyloid protein (YENPTY) contains an NPXY motif (18). However, tyrosine phosphorylation of this site is not required for binding, and indeed the binding affinity is not compromised in in vitro binding assays even when the tyrosine residue in the NPTY sequence is mutated (18). Furthermore, the PTB domains from FE65 and X11 apparently recognize different sites in the YENPTY sequence, as suggested by mutagenesis studies (18). These studies raise the possibility that the PTB domain may represent a family of protein modules with diverse ligand-binding specificity that is not necessarily restricted to recognition of the NPXpY motif (19).
The membrane-associated protein Numb also contains a PTB domain whose specificity and biochemical function has not been well characterized. Numb was originally identified as a gene required for specifying cell type during the development of the Drosophila peripheral nervous system (20). The Drosophila numb gene encodes a 61-kDa protein, dNumb, which is segregated selectively into one daughter cell during the binary division of the sensory organ precursor (SOP), and functions as an intrinsic determinant of cell fate by controlling asymmetric cell division (20, 21). Part of the N-terminal portion of the dNumb protein exhibits sequence similarity to the Shc PTB domain, suggesting that dNumb may contain a functional PTB domain (10). Genetically, dNumb appears to act as an inhibitor of the cell-surface receptor Notch, which may provide a mechanism through which dNumb induces a neuronal phenotype (22). It was recently suggested that the PTB domain interacts with the RAM-23 region of the proneural protein Notch, and thereby mediates a physical association between these two proteins (22, 23). However, the PTB-binding motif in Notch has not been determined. We have undertaken a different approach to investigate the specificity of the dNumb PTB domain. By expressing the dNumb PTB domain as a GST fusion protein and studying its binding to a Tyr-oriented synthetic peptide library, we have identified a novel peptide motif, GP(p)Y, that is specifically recognized by the PTB domain of dNumb. The specificity of the dNumb PTB domain was further explored using synthetic peptide analogues and spectroscopic methods, including surface plasmon resonance (SPR), fluorescence polarization, and NMR. Our results suggest that the dNumb PTB domain possesses a unique binding specificity that is distinct from the motifs recognized by other known PTB domains.
MATERIALS AND METHODS
Peptide Synthesis and Purification.
A peptide library containing the generic sequence H2N-MAXXXX-Y-XXXX AKKK-OH (X = any amino acid; Y = tyrosine) was generated following published procedures (11). Individual peptides were synthesized and purified following published procedures (12). The identities of the peptides were confirmed by mass spectrometry and amino acid analysis. The concentrations of peptide stock solutions were determined by measuring the absorbance at 264 nm using an extinction coefficient of 1,752 M−1cm−1 for pTyr and 840 M−1cm−1 for Tyr (24).
Expression and Purification of the PTB Domain of dNumb.
The dNumb PTB domain (residues 58–205) was subcloned into a pGEX4T2 vector using the restriction endonuclease sites BamHI (5′) and XhoI (3′). The GST-fused dNumb PTB domain was expressed in bacteria BL21 DE3 cells and purified following published procedures (12). The isolated PTB domain was generated by thrombin cleavage, followed by purification on a Superdex G-75 column (Pharmacia) (12).
SPR Analysis of PTB–Peptide Interaction.
Surface plasmon resonance measurements were conducted on a BIAcore apparatus (Pharmacia Biosensor). Peptides were blocked at the N terminus by acetylation. Peptides used for immobilization also contained a GGK extension at the C terminus to facilitate their coupling onto the CM5-type Biosensor chip. The GGK extension should also alleviate constraints to the peptide conformation that might result from immobilization. Sensorgrams of protein–peptide interactions and peptide competition were recorded using similar procedures as reported previously (12).
Fluorescence Polarization.
Fluorescence polarization experiments were conducted on a Beacon Fluorescence Polarization System (PanVera, Madison, WI) equipped with a 100-μl sample chamber. Peptides containing a C-terminal GGK extension were used for fluorescence labeling with 6-(fluorescein-5-(and-6)-carboxamido)hexanoic acid, succinimidyl ester (5(6)-SFX) (Molecular Probes). For fluorescence polarization studies, the labeled peptides were dissolved in 20 mM phosphate, pH 6.0, containing 100 mM NaCl, 0.1 mM EDTA, 2 mM DTT, and 0.1 mM benzamidine, and diluted to give a polarization value of 50–70 mP. Buffers and solutions used in the experiments were either filtered through a 0.22-μm filter or spun at 10,000 rpm for 5 min to remove particulates. For competition studies, various concentrations of unlabeled peptides were mixed with 5.0 μM protein in the presence of labeled peptides, and the mixture was allowed to stand at room temperature for 5 min before measurement. All measurements were carried out at 25°C.
NMR Spectroscopy.
15N,13C-labeled protein samples were obtained by growing the transformed bacteria in a minimal medium containing 15NH4Cl and 13C-labeled glucose as the sole sources of nitrogen and carbon. Purification of the protein followed the same procedure as described above. NMR experiments were performed on Varian Unity 500- and 600-MHz spectrometers equipped with actively shielded Z-gradient probes and gradient amplifier units. The buffer used in NMR studies contained 20 mM phosphate (pH 6.0), 100 mM NaCl, and 0.1 mM perdeuterated EDTA in H2O/D2O (9/1) (Cambridge Isotope Laboratories, Cambridge, MA). Titration of the protein with the desired peptide dissolved in the same buffer was monitored by changes in 15N-1H heteronuclear single-quantum correlation (HSQC) spectra (25). For backbone assignment of the protein, triple resonance experiments including HNCO, HNCACB, HBCBCA(CO)NNH, HBHA(CO)CAHA, and CCC-TOCSY-(CO)NNH (25) were recorded. The data were processed using the nmrpipe/nmrdraw package (26) and analyzed using nmrview3 (27).
RESULTS
Identification of a dNumb PTB-Binding Motif.
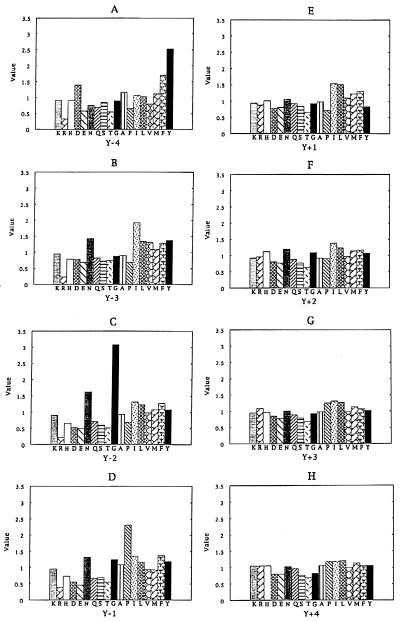
A Tyr-oriented peptide library containing the degenerate amino acid sequence of XXXX-Y-XXXX (where X represents any naturally occurring amino acid except Cys and Trp) was synthesized to search for peptides that interact with the PTB domain of the Drosophila Numb protein. The dNumb PTB domain, expressed in Escherichia coli as a GST fusion protein, was purified on glutathione-agarose beads and used to screen against the peptide mixture from the library. The bound peptides were then eluted, pooled, and sequenced. Selectivity at each of the degenerate positions was determined by comparing the abundance of individual amino acids in the eluate with the corresponding abundance in a control experiment where GST-containing beads were used (11). The results obtained are shown in Fig. 1.
Figure 1.
Selection of the dNumb PTB domain for individual amino acids at the “X” positions in the degenerate library MAXXXX-Y-XXXXAKKK. The frequency of occurrence at each position was calculated according to the amino acid’s abundance in the bound mixture relative to its abundance in a control experiment.
Strong selection was observed for residues located N-terminal to the fixed Tyr, whereas selection at the C-terminal positions was largely insignificant. In particular, aromatic residues such as Tyr and Phe were favored at position Y-4, and the hydrophobic residue Ile was slightly preferred to other amino acids at position Y-3. However, the strongest selection was observed for position Y-2 (Fig. 1C), which was dominated by Gly, although the small residue Asn was also selected. A Pro residue was strongly selected at position Y-1. In contrast, a relatively weak bias for hydrophobic residues (e.g., Leu, Ile, Phe) was seen for position Y+1, whereas positions Y+2, Y+3, and Y+4 did not seem to favor any particular residue type (Fig. 1 E–H). Taken together, these data predict that the dNumb PTB domain binds preferentially to peptides containing the sequence motif YIGPYφ (φ denotes a hydrophobic residue).
A Peptide Containing the YIGPYφ Motif Binds Specifically to the Isolated dNumb PTB Domain.
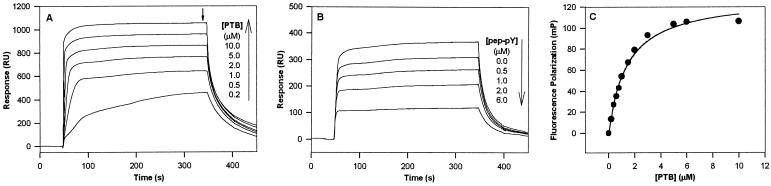
To test the binding of the selected peptide sequence to the dNumb PTB domain, we synthesized a peptide containing the motif YIGPYφ. The peptide had the sequence Ac-AYIGPYL-OH and was designated pep-Y. A modified version of pep-Y was also synthesized with a -GGK sequence added to the C terminus to create a flexible anchoring site for plasmon resonance and fluorescence polarization studies. Peptide pep-Y (GGK) was readily immobilized on a CM5 sensor chip via the ɛ-NH2 of the Lys side chain by amine coupling. Interaction of the dNumb PTB domain with pep-Y (GGK) was then examined by recording the corresponding SPR sensorgrams following the injection of various concentrations of isolated protein samples across the chip surface. As shown in Fig. 2A, the dNumb PTB domain bound to the immobilized peptide pep-Y in a concentration-dependent manner. Strong binding signals were generated when micromolar concentrations of the dNumb PTB protein were applied. The dissociation constant of the dNumb PTB/pep-Y complex was estimated to be 8.07 ± 0.96 μM from a hyperbolic fitting of the equilibrium SPR responses at various protein concentrations (Fig. 2A; calculation not shown). To examine the specificity of the pep-Y–dNumb PTB interaction, various concentrations of free peptide pep-Y were mixed with 5.0 μM dNumb PTB protein prior to sample application. As illustrated in Fig. 2B, the signal resulting from dNumb PTB binding to immobilized pep-Y(GGK) could be competed away when free peptide pep-Y was added to the buffer, suggesting that the dNumb PTB-pep-Y interaction is specific.
Figure 2.
(A) SPR response of dNumb PTB binding to immobilized pep-Y. Concentrations of the dNumb PTB protein were as indicated. The arrow indicates the time where points were taken for the estimation of Kd from a hyperbolic fitting of the binding curve. RU, resonance unit. (B) Inhibition of dNumb PTB binding to immobilized pep-Y by soluble pep-Y added into the medium. PTB domain concentration used was 5.0 μM. Concentrations of the competing peptides were as indicated. (C) Binding of the isolated PTB domain of dNumb to fluorescein-labeled pep-Y. Fluorescence polarization values (mP) reflect the difference in polarization before and after the PTB domain was added to the medium. Parallel experiments generally gave polarization values with deviations under 10% of the reported values.
To investigate whether there is a direct binding of the dNumb PTB protein with peptide pep-Y in solution, pep-Y (GGK) was covalently linked to fluorescein via its Lys side chain ɛ-NH2 group. The peptide–protein interaction was then studied by fluorescence polarization. Because fluorescence polarization is directly related to the size of the molecules to which the fluorophore is attached, the binding affinity of pep-Y to the dNumb PTB domain could be readily determined by monitoring the level of fluorescence polarization following the addition of incremental amounts of isolated dNumb PTB protein. As can be appreciated from Fig. 2C, the interaction between pep-Y and dNumb PTB was concentration-dependent and followed a saturable pattern. Hyperbolic fitting of the binding data yielded a dissociation constant of 5.78 ± 0.74 μM for the complex, which agrees essentially with the result from SPR studies.
Tyrosine Phosphorylation Enhances Binding of pep-Y to the dNumb PTB Domain.
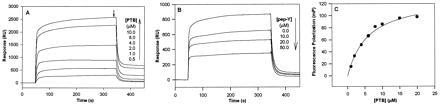
A phosphorylated version of pep-Y, Ac-AYIGPpYL-OH (designated pep-pY), was also synthesized to explore the effect of tyrosine phosphorylation on peptide–dNumb PTB binding. Because the YIGPYφ motif was identified from a Tyr-oriented library screening, one might expect that pep-pY would not be a good ligand for the dNumb PTB domain. Interestingly, however, peptide pep-pY bound with high affinity to the dNumb PTB domain. SPR studies employing a sensor chip coupled with pep-pY showed that the PTB domain of dNumb bound to phosphorylated pep-pY in a similar fashion as to unphosphorylated pep-Y (Fig. 3A). However, less protein was needed to saturate the SPR signal as the dissociation constant (Kd) of the dNumb PTB/pep-pY complex was reduced to 0.69 ± 0.11 μM, as estimated from hyperbolic fitting of the equilibrium binding signal (Fig. 3A; calculation not shown). Similarly, the dNumb PTB–pep-pY interaction was shown to be specific, because free pep-pY could inhibit dNumb PTB binding to immobilized pep-pY (Fig. 3B). A control phosphopeptide, IGVPVSVDNPEpYLLNAQK, derived from the dShc PTB-binding site in the Drosophila EGF receptor (DER) tyrosine kinase (12), was immobilized on a sensor chip to test its binding to the dNumb PTB domain. No interaction was observed when the dNumb PTB protein was applied at 100 μM on the chip immobilized with the DER phosphopeptide (data not shown). We also tested the binding of the dShc PTB domain to immobilized pep-pY; no binding signal was detected in the corresponding SPR sensorgram (data not shown). These studies suggest that the PTB domains from dShc and dNumb have very different binding specificities toward phosphopeptides.
Figure 3.
(A) SPR response of dNumb PTB binding to immobilized pep-pY. Concentrations of the dNumb PTB protein were as indicated. (B) Inhibition of dNumb PTB binding to immobilized pep-pY by soluble pep-pY. PTB concentration was 1.0 μM. Concentrations of the peptide were as indicated. (C) Binding of the isolated PTB domain of dNumb to fluorescein-labeled pep-pY.
Fluorescence polarization was also employed to examine the pep-pY–dNumb PTB interaction in solution. As shown in Fig. 3C, the dNumb PTB domain binds to fluorescein-labeled pep-pY in a concentration-dependent manner with a dissociation constant of 1.41 ± 0.10 μM. This value is slightly higher than that obtained from SPR analysis. Nevertheless, phosphorylation of the second Tyr residue in the YIGPYφ motif yielded a peptide whose affinity for the dNumb PTB domain was significantly enhanced compared with pep-Y. It is interesting to note that the affinity of the dNumb PTB domain for phosphorylated pep-pY is comparable to that of the Shc PTB domain for its phosphopeptide ligands (28), and is higher than the reported affinity of the PTB domain of IRS-1 for phosphopeptides derived from the PTB-binding sites in the IR (28).
The Core Sequence, GPpY, Directs High-Affinity Binding of Pep-pY to the dNumb PTB Domain.
To identify key residues within the YIGPpYL sequence responsible for high-affinity binding of pep-pY to the dNumb PTB domain, we synthesized a series of peptides, each of which contained a single residue substitution (Table 1). The relative affinities of these variant peptides for the dNumb PTB domain were measured by their ability to inhibit the binding of the dNumb PTB domain to fluorescein-labeled pep-Y by using fluorescence polarization. The IC50 values for each peptide were determined and compared with that of pep-pY to yield their relative binding affinities. Peptide pep-Y inhibited its own binding to dNumb PTB with an IC50 value of ca. 15.6 μM, whereas pep-pY inhibited with a much lower IC50 of ca. 1.74 μM (Table 1). Phosphorylation of the first Tyr residue in the YIGPYL sequence produced peptide Y−4-pY, whose affinity to the dNumb PTB domain was markedly reduced compared with pep-pY, suggesting that the phosphorylation of the second Tyr rather than the first Tyr residue in the YIGPYL motif can promote dNumb PTB binding. Substituting residue Tyr−4 with Ala in pep-pY resulted in an approximately 8-fold loss of binding affinity (Table 1), indicating that Tyr−4 plays a significant role in pep-pY–dNumb PTB interaction. In contrast, replacement of the bulky, hydrophobic residue Ile−3 with the small, yet hydrophobic amino acid Ala had a less drastic effect, as the resulting peptide I−3-A retained approximately 30% of the affinity observed for pep-pY. Thus, a hydrophobic residue at the Y-3 position may not be crucial for binding. However, substitution of either Gly−2, Pro−1, or pTyr0 by Ala almost completely abolished the binding activity of the corresponding peptides (G−1-A, P−1-A, and pY0-A), suggesting that these three residues are critical for high-affinity dNumb PTB binding. The bulky, hydrophobic residue, Leu, at the Y+1 position appears not to be required for binding, because a peptide with the Ala at +1 position retained full binding affinity. Interestingly, an Asn-for-Gly substitution at position −2 had a less detrimental effect on dNumb PTB binding than the corresponding Ala-for-Gly substitution, whereas replacing Pro−1 with either Asn or Ala had the same inhibitory effect. The relatively high affinity observed for peptide G−1-N appears to be in agreement with the data from peptide library screening, in that Asn was identified as the second most strongly selected residue at position Tyr-1 (Fig. 1). In control experiments, a Shc PTB-binding peptide derived from the TrkA receptor (either phosphorylated or unphosphorylated on Tyr) (12, 14), and two IRS-1 PTB-binding peptides derived from the IR and the IL4 receptor (IL4R), respectively (12, 15, 16), failed to inhibit the dNumb PTB–pep-Y interaction at 1.0 mM. Therefore, it is likely that the sequence GP(p)Y functions as a core motif that mediates specific and high-affinity binding of peptide pep-Y/pep-pY to the dNumb PTB domain.
Table 1.
Relative affinity of peptides to the dNumb PTB domain measured in solution
| Peptide | Amino acid sequence | IC50 ± SE, μM* | Relative affinity, %† |
|---|---|---|---|
| −4 −3 −2 −1 0 +1 | |||
| pep-Y | Ac-A Y I G P Y L-OH | 15.6 ± 1.6 | 11.1 |
| pep-pY | Ac-A Y I G P pY L-OH | 1.74 ± 0.20 | 100 |
| Y−4-pY | Ac-A pY I G P Y L-OH | 12.6 ± 0.9 | 13.8 |
| Y−4-A | Ac-A AI G P pY L-OH | 12.5 ± 0.6 | 13.9 |
| I−3-A | Ac-A YA G P pY L-OH | 6.0 ± 0.6 | 29.0 |
| G−2-A | Ac-A YI A P pY L-OH | 500 ± 65 | 0.3 |
| G−2-N | Ac-A YI N P pY L-OH | 8.1 ± 1.2 | 21.5 |
| P−1-A | Ac-A YI G A pY L-OH | 79.4 ± 5.3 | 2.2 |
| P−1-N | Ac-A YI G N pY L-OH | 112 ± 7 | 1.6 |
| pY0-A | Ac-A Y I G P A L-OH | 56.2 ± 6.5 | 3.1 |
| L+1-A | Ac-A YI G P pY A-OH | 1.81 ± 0.44 | 96.7 |
| TrkA (pY490) | H I I E N P G pY F S D | >1,000 | ND |
| TrkA (Y490) | H I I E N P G Y F S D | >1,000 | ND |
| IR (pY960) | Y A S S N P E pY L S A | >1,000 | ND |
| IL4R (pY475) | V L A D N P A pY R S F | >1,000 | ND |
IC50 peptide concentration required to achieve 50% inhibition of dNumb PTB binding to fluorescein-labeled peptide pep-Y. Reported values with the calculated standard errors (SE) were based on three parallel experiments.
Relative affinity of the peptides for dNumb PTB domain calculated according to their IC50 values. The affinity of pep-pY was assigned as 100%. ND, not determined.
NMR Spectroscopy Shows That the dNumb PTB Domain Binds to Both Phosphorylated and Nonphosphorylated Sequences.
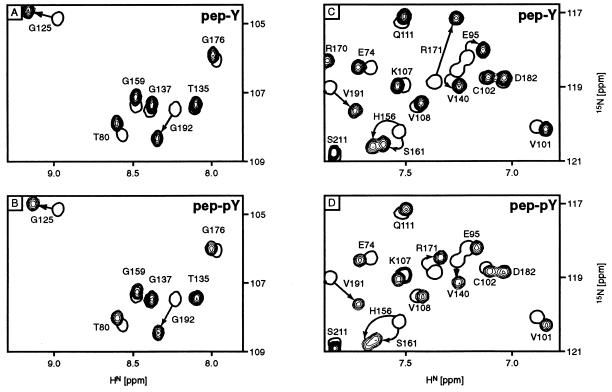
The protein backbone amide 1H-15N peaks in an HSQC spectrum are sensitive to changes in the environment and can be used to monitor protein–ligand interactions. The 1H-15N HSQC spectrum recorded for the free dNumb PTB domain in phosphate buffer, pH 6.0, displayed good dispersion of the peaks over a wide range of amide proton chemical shifts from 6.0 to 10.3 ppm, indicating that the protein is folded under these experimental conditions. Addition of peptide pep-Y in a stepwise fashion into the PTB sample to a molar ratio of 1:1 induced significant shifts of many of the peaks in the HSQC spectrum, demonstrating an interaction between the dNumb PTB protein and the peptide. Interestingly, the pTyr-containing peptide pep-pY was found to induce similar patterns of chemical shift changes in the HSQC spectra in a parallel peptide titration experiment. Two representative sections of the HSQC spectra displaying the chemical shift changes resulting from protein–peptide complex formation are shown in Fig. 4. It is evident from a comparison of Fig. 4A with 4B and Fig. 4C with 4D that the binding of pep-Y and pep-pY to the dNumb PTB domain involves the same set of amino acids in the protein. The largest shifts, including those of resonances from residues G125 and G192 (Fig. 4 A and B), H156, S161, R171, and V191 (Fig. 4 C and D), are suggestive of either direct contacts between these amino acids and the peptide or structural perturbation of these residues due to ligand binding. Residue V191 of the dNumb PTB domain corresponds to I194 in the Shc PTB domain, which participates in hydrophobic interactions with the hydrophobic side chain of Ile(pY-5) of the TrkA peptide in the Shc PTB/TrkA complex (14). Residue G192 of Numb corresponds to the immediately adjacent G195 in Shc, a Gly that is conserved in all known PTB domains and is also located at the peptide-binding interface. Similarly, dNumb R171 corresponds to R175 in the Shc PTB domain, a residue that is conserved among PTB domains and involved in critical interactions with the phosphotyrosine in the TrkA complex. Of note, mutation of this residue in Shc abrogates TrkA binding (7, 14). Interestingly, the extremely large 15N chemical shift change for R171 upon pep-Y binding is significantly reduced in the pep-pY complex. This is one of the most notable differences in the chemical shift patterns upon titration with the two peptides. The large upfield shift in the pep-Y complex is consistent with a ring-current shift due to placement of the 15N above or below the second tyrosine ring of the peptide. Presence of a phosphate may lead to downfield shifts, depending on the exact geometry, and these opposing effects may explain the reduction in the chemical shift change in the pep-pY complex. Other chemical shift changes, including those for residues G125, H156, and S161, are not as easily explained by direct peptide interactions in the homologous Shc PTB/TrkA complex. These shifts may be due to indirect structural perturbation. Overall, these NMR titration data demonstrate that peptides pep-Y and pep-pY recognize the same set of residues in the dNumb PTB domain. In addition, a number of residues previously shown to be critical for the Shc PTB domain–peptide ligand interaction also appear to play important roles in the dNumb PTB–pep-Y/pep-pY interaction. Further studies of the three-dimensional structure of the dNumb PTB–pep-Y/pep-pY complexes should shed light on differences in specificity between the Shc and dNumb PTB domains.
Figure 4.
Overlay plots of HSQC spectra for the free dNumb PTB domain (open), and the PTB domain bound to unphosphorylated pep-Y (A and C) and phosphorylated pep-pY (B and D) (solid). For clarity, only small sections of the HN-15N region are shown. Resonance peaks are labeled according to their numbering in the original protein sequence (20). Peaks displaying large chemical shift changes between the free and complexed states are identified by arrows.
DISCUSSION
Using a degenerate peptide library MAXXXX-Y-XXXXAKKK, where a Tyr residue is held constant in the middle of the peptide sequence, we have identified a YIGPYφ motif that interacts specifically with the dNumb PTB domain. Peptides containing this motif bind with high affinity to the isolated PTB domain of dNumb in solution as well as on a BIAcore. Like other PTB domains, the selectivity of the dNumb PTB domain is primarily confined to residues N-terminal to the fixed Tyr. However, the motif selected by the dNumb PTB domain is markedly different from those recognized by the PTB domains of Shc and IRS-1, which preferably bind to peptides containing an NPXpY motif. Moreover, phosphorylation of the Tyr residue within the NPXY motif is indispensable for binding to Shc and IRS-1 PTB domains, because nonphosphotylated peptides generally display at least a thousand-fold decrease in affinity (28). In contrast, dephosphorylation of the second Tyr within the YIGPpY motif reduced its binding affinity for the dNumb PTB domain by less than 10-fold. Thus, the dNumb PTB domain is able to bind to both Tyr- and pTyr-containing peptides with dissociation constants in the micromolar range. NMR studies on the complexes formed by pep-Y and pep-pY with the isolated PTB domain of dNumb suggest that the two peptides interact with the same binding site in the protein. Structural studies of these complexes by NMR should yield useful information about the mechanism by which the dNumb PTB domain can recognize both Tyr- and pTyr-containing sequences. The role played by individual residues within the YIGP(p)Yφ motif on dNumb PTB binding has been explored by substitution studies. In agreement with the library screening results, a core segment, GPpY, appears to be essential for high-affinity dNumb PTB-binding, whereas residues such as Y−4 and I−3 likely play a more minor role. It is interesting to note that the PTB domains from two other proteins, X11 and FE65, also recognize a Tyr-containing motif (YENPTY), which superficially resembles the YIGP(p)Y sequence recognized by the dNumb PTB domain. Nevertheless, it was shown previously that the dNumb protein is unable to bind to the YENPTY sequence (23).
The PTB domains from Shc and IRS-1 are known to require hydrophobic residues N-terminal to the NPXpY motif for high-affinity binding. Does the dNumb PTB domain also recognize residues beyond the YIGP(p)Yφ motif? Although this question has not been explicitly addressed in the current study, a peptide, H2N-AYIGP(p)YL-OH, with its N terminus left unblocked, exhibited significantly reduced affinity for the dNumb PTB domain compared with either the acetylated pep-Y or pep-pY (data not shown). Whether the dNumb PTB domain prefers hydrophobic residue(s) at positions upstream of Tyr-4 awaits further investigation.
Defining the structural basis for the specificity of the dNumb PTB domain will require solving the structure of the dNumb PTB domain in complex with the peptide pepY or pep-pY. However, NMR assignment of the resonances of the protein has already demonstrated that the dNumb PTB domain has a secondary structure similar to the Shc PTB domain (S.J.F.V., C.Z., and S.-C.L., unpublished results). It is likely that the PTB domains from different proteins have a similar general fold, as has been shown for the PTB domains of Shc and IRS-1 (14–16). The diversity in specificity then must come from differences in the binding sites. Indeed, the Shc PTB domain and the IRS-1 PTB domain employ distinct sets of basic residues in pTyr binding, although their three-dimensional structures are very similar. It is conceivable that the GP(p)Y motif recognized by the dNumb PTB domain may also adopt a turn structure similar to the β-turn formed by the NPXpY motif upon its binding to the PTB domains of Shc and IRS-1. In support of this notion, residues such as Gly, Pro, and Asn are optimal amino acids for the formation of β-turns in proteins (29). However, that the dNumb PTB domain binds specifically to a GP(p)Y rather than an NPXpY sequence suggests that the structure adopted by the two motifs may differ in detail.
In summary, we have identified a novel peptide motif, GP(p)Y, that is recognized specifically by the dNumb PTB domain. Although we do not know whether this represents a physiological binding site and we cannot exclude the possibility of alternative Numb PTB-binding sequences, our finding that the dNumb PTB domain possesses a unique specificity toward Tyr- and pTyr-containing peptide sequences demonstrates that PTB domains in general may recognize a diverse array of peptide sequences that likely form turn structures, but are not necessarily phosphorylated. Such variation in peptide recognition by PTB domains presumably reflects the wide range of biological activities undertaken by PTB-containing proteins. In some instances PTB domains may also bind specific phospholipids (14), an activity that is also demonstrated by PH domains which possess a fold very similar to PTB domains (14, 30). It is of considerable interest that a protein module originally identified through its role in tyrosine kinase signaling is also found in a protein that controls asymmetric cell division. In this regard, it is intriguing that the localization of dNumb is itself regulated by another protein, Inscutable, that also specifies the orientation of mitotic spindle formation (31). Inscutable possesses multiple ankyrin repeats and potential binding sites for WW domains (PPPPY) and PDZ domains (ESDV-COOH) (32). These observations attest to the broad array of cellular processes that make use of modular protein–protein interactions.
Acknowledgments
We thank Dr. Y. N. Jan for kindly providing the Drosophila Numb cDNA. S.-C.L. is a postdoctoral fellow of the Medical Research Council of Canada. S.V. is a postdoctoral fellow of the Swiss Science National Fund. C.Z. is the recipient of a Swiss Science National Fund fellowship and a Human Frontier Science Program fellowship. This work was supported by grants from the Human Frontier Science Program, Asahi Chemical Company, the National Cancer Institute of Canada (NCIC), and the Medical Research Council of Canada, and by a Howard Hughes International Research Scholar award to T.P. T.P. is a Terry Fox Cancer Research Scientist of the NCIC. L.E.K. and J.F.-K. acknowledge financial support from the NCIC.
ABBREVIATIONS
- PTB
phosphotyrosine binding
- NMR
nuclear magnetic resonance
- PI
phosphotyrosine interaction
- IR
insulin receptor
- SPR
surface plasmon resonance
- HSQC
heteronuclear single-quantum correlation
References
- 1.Hunter T. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 2.Pawson T. Nature (London) 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 3.Cohen G B, Ren R, Baltimore D. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 4.Kavanaugh W M, Williams L T. Science. 1994;266:1862–1865. doi: 10.1126/science.7527937. [DOI] [PubMed] [Google Scholar]
- 5.Blaikie P, Immanuel D, Wu J, Li N, Yajnik V, Margolis B. J Biol Chem. 1994;269:32031–32034. [PubMed] [Google Scholar]
- 6.Van der Geer P, Wiley S, Lai V K-M, Olivier J P, Gish G D, Stephens R, Kaplan D, Shoelson S E, Pawson T. Curr Biol. 1995;5:404–412. doi: 10.1016/s0960-9822(95)00081-9. [DOI] [PubMed] [Google Scholar]
- 7.Van der Geer P, Wiley S, Gish G, Lai V K-M, Stephens R, White M F, Pawson T. Proc Natl Acad Sci USA. 1996;93:963–968. doi: 10.1073/pnas.93.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun X J, Wang L-M, Zhang Y, Yenush L, Myers M G, Glasheen E, Lane S, Pierce J H, White M F. Nature (London) 1995;377:173–177. doi: 10.1038/377173a0. [DOI] [PubMed] [Google Scholar]
- 9.Sawka-Verhelle D, Tartare-Dechert S, White M S, Van Obberghen E. J Biol Chem. 1996;271:5980–5983. doi: 10.1074/jbc.271.11.5980. [DOI] [PubMed] [Google Scholar]
- 10.Bork J-P, Margolis B. Cell. 1995;80:693–694. doi: 10.1016/0092-8674(95)90347-x. [DOI] [PubMed] [Google Scholar]
- 11.Songyang Z, Shoelson S E, Chaudhuri M, Gish G, Pawson T, Haser W G, King F, Roberts T, Ratnofsky S, Lechleider R J, Neel B G, Birge R B, Fajardo J E, Chou F M, Hanafusa H, Schaffhausen B, Cantley L C. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 12.Li S-C, Lai V K-M, Gish G D, Parris W, van der Geer P, Forman-Kay J, Pawson T. J Biol Chem. 1996;271:31855–31862. doi: 10.1074/jbc.271.50.31855. [DOI] [PubMed] [Google Scholar]
- 13.Wolf G, Trub T, Ottinger E, Groninga L, Lynch A, White M, Shoelson S. J Biol Chem. 1995;270:27407–27410. doi: 10.1074/jbc.270.46.27407. [DOI] [PubMed] [Google Scholar]
- 14.Zhou M-M, Ravichandran K S, Olejniczak E T, Petros A M, Meadows R P, Sattler M, Harlan J E, Wade W S, Burakoff S J, Fesik S W. Nature (London) 1995;378:584–592. doi: 10.1038/378584a0. [DOI] [PubMed] [Google Scholar]
- 15.Eck M J, Dhe-Paganon S, Trub T, Nolte R, Shoelson S E. Cell. 1996;85:695–705. doi: 10.1016/s0092-8674(00)81236-2. [DOI] [PubMed] [Google Scholar]
- 16.Zhou M-M, Huang B, Olejniczak E T, Petros A M, Meadows R P, Shuker S B, Miyazaki M, Trub T, Fesik S W. Nat Struct Biol. 1996;3:388–393. doi: 10.1038/nsb0496-388. [DOI] [PubMed] [Google Scholar]
- 17.Guenette S Y, Chen J, Jondro P D, Tanzi R E. Proc Natl Acad Sci USA. 1996;93:10832–10837. doi: 10.1073/pnas.93.20.10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borg J-P, Ooi J, Levy E, Margolis B. Mol Cell Biol. 1996;16:6229–6241. doi: 10.1128/mcb.16.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charest A, Wagner J, Jacob S, McGlade C J, Tremblay M L. J Biol Chem. 1996;271:8424–8429. doi: 10.1074/jbc.271.14.8424. [DOI] [PubMed] [Google Scholar]
- 20.Uemura T, Shephard S, Ackerman L, Jan L Y, Jan Y N. Cell. 1989;58:349–360. doi: 10.1016/0092-8674(89)90849-0. [DOI] [PubMed] [Google Scholar]
- 21.Rhyu M S, Jan L Y, Jan Y N. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 22.Frise E, Knoblich J A, Younger-Shepherd S, Jan L Y, Jan Y N. Proc Natl Acad Sci USA. 1996;93:11925–11932. doi: 10.1073/pnas.93.21.11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo M, Jan L Y, Jan Y N. Neuron. 1996;17:27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 24.Turck C W. Pept Res. 1992;5:156–160. [PubMed] [Google Scholar]
- 25.Cavanagh J, Fairbrother W J, Palmer A G, III, Skelton N J. Protein NMR Spectroscopy, Principles and Practice. San Diego: Academic; 1996. [Google Scholar]
- 26.Delaglio F, Grzesiek S, Vuister G W, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 27.Johnson B A, Blevins R A. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 28.Zhou M-M, Harlan J E, Wade W S, Crosby S, Ravichandran K S J, Burakoff J, Fesik S W. J Biol Chem. 1995;270:31119–31123. doi: 10.1074/jbc.270.52.31119. [DOI] [PubMed] [Google Scholar]
- 29.Chou P Y, Fasman G D. J Mol Biol. 1977;115:135–175. doi: 10.1016/0022-2836(77)90094-8. [DOI] [PubMed] [Google Scholar]
- 30.Harlan J E, Hajduk P J, Yoon H-S, Fesik S W. Nature (London) 1994;371:168–170. doi: 10.1038/371168a0. [DOI] [PubMed] [Google Scholar]
- 31.Kraut R, Chia W, Jan L Y, Jan Y N, Knoblich J A. Nature (London) 1996;383:50–55. doi: 10.1038/383050a0. [DOI] [PubMed] [Google Scholar]
- 32.Kraut R, Campos-Ortega J A. Dev Biol. 1996;174:65–81. doi: 10.1006/dbio.1996.0052. [DOI] [PubMed] [Google Scholar]