Abstract
Regulators of G protein signaling (RGS) proteins act as GTPase-activating proteins (GAPs) toward the α subunits of heterotrimeric, signal-transducing G proteins. RGS11 contains a G protein γ subunit-like (GGL) domain between its Dishevelled/Egl-10/Pleckstrin and RGS domains. GGL domains are also found in RGS6, RGS7, RGS9, and the Caenorhabditis elegans protein EGL-10. Coexpression of RGS11 with different Gβ subunits reveals specific interaction between RGS11 and Gβ5. The expression of mRNA for RGS11 and Gβ5 in human tissues overlaps. The Gβ5/RGS11 heterodimer acts as a GAP on Gαo, apparently selectively. RGS proteins that contain GGL domains appear to act as GAPs for Gα proteins and form complexes with specific Gβ subunits, adding to the combinatorial complexity of G protein-mediated signaling pathways.
Proteins belonging to the RGS (regulators of G protein signaling) family constitute a newly appreciated group of at least 20 mammalian gene products that act as GTPase-activating proteins (GAPs) on the α subunits of heterotrimeric, signal-transducing G proteins (1–3). As such, RGS proteins can serve as negative regulators of G protein-mediated signaling pathways by speeding the inactivation of GTP-bound Gα subunits. Although several members of the RGS family are relatively simple ≈25 kDa proteins that contain little more than a characteristic RGS domain, others include modules that impart additional functions. For example, RGS12 can associate in vitro with certain G protein-coupled receptors by virtue of an alternatively spliced PDZ (PSD-95/Dlg/Z0-1) domain (4), and p115, a guanine nucleotide exchange factor for the low-molecular-weight GTPase rho, contains an RGS domain that imparts sensitivity to regulation by G protein α subunits (5, 6).
We describe here a novel G protein γ subunit-like domain (GGL; pronounced giggle) that is found in several mammalian RGS proteins (RGS6, RGS7, RGS9, and RGS11) and in EGL-10, an RGS protein of Caenorhabditis elegans. The GGL domains of RGS11 and RGS7 interact preferentially with the G protein β5 subunit, and the complex of RGS11 and β5 has GAP activity toward the G protein αo subunit.
MATERIALS AND METHODS
Generation of Expression Constructs.
cDNAs for RGS11 and various G protein subunits were cloned from human brain or retinal mRNA, from mouse retinal mRNA, or were obtained as described (7, 8); all amplified cDNAs were verified by sequencing. Human RGS7 cDNA was a kind gift of Paul F. Worley (Johns Hopkins University). cDNAs encoding G protein subunits were subcloned into the mammalian expression vector pcDNA3.1-Zeo (Invitrogen), and Gγ and RGS protein cDNAs were subcloned in-frame with an N-terminal tandem hemagglutinin (HA)-epitope tag into a modified pcDNA3.1 vector. Recombinant baculoviruses expressing native or hexahistidine-tagged RGS11 or Gβ5 subunits were generated by using the Bac-To-Bac system by following the manufacturer’s protocols (Life Technologies, Gaithersburg, MD).
In Vitro Transcription and Translation.
Reactions were performed using the TNT reticulocyte lysate system (Promega), with conditions essentially as described by Schmidt and Neer (9). Reaction mixtures were incubated at 30°C for 1 hr; appropriate reactions were then combined and allowed to transcribe/translate for an additional 1 hr at 37°C before immunoprecipitation in the presence of 0.05% C12E10, 20% glycerol, and protease inhibitors by using protein A-Sepharose-CL4B (Sigma) and anti-HA mAb 12CA5 (Boehringer Mannheim). Protein A beads were washed, suspended in 2× Laemmli sample buffer, and boiled for 5 min. Proteins were separated by SDS/PAGE on Tris-glycine gels.
Anti-RGS11 Antibody.
A cDNA fragment encoding the RGS11 GGL domain (aa 219–292) was subcloned into the glutathione S-transferase fusion (GST) vector pGEX4T3 (Pharmacia), and fusion protein was expressed and purified as described (4). Purified protein and complete Freund’s adjuvant were injected into New Zealand White rabbits (Antibodies Inc.). Crude antisera were depleted with GST-coupled CNBr-Sepharose and affinity purified with GST-RGS11 (aa 219–467)-coupled CNBr-Sepharose. Antibody elution was performed with 0.1 M glycine, pH 2.5.
Transient Transfection and Immunoprecipitation.
COS-7 cells were transfected with SuperFect reagent and DNA from each expression plasmid by following the manufacturer’s instructions (Qiagen, Chatsworth, CA). Cells were harvested 48 hr later by scraping into 1 ml of RIPA-150 buffer (150 mM NaCl/50 mM Tris⋅HCl, pH 7.5/20 mM EDTA/0.5% deoxycholate/1% Nonidet P-40/0.1% SDS/protease inhibitors), lysed, and clarified by centrifugation. Supernatants were cleared for 30 min at 4°C with 50% (vol/vol) protein A-Sepharose, incubated with 12CA5 antibody for 1 hr at 4°C, and then cleared with 50% (vol/vol) protein A-Sepharose for an additional hour. Protein A beads were washed three times in RIPA-150 buffer, suspended in 2× Laemmli sample buffer (NOVEX), and boiled for 5 min. Proteins were separated by SDS/PAGE, electroblotted onto nitrocellulose, and detected with appropriate antisera, secondary antibodies conjugated to horseradish peroxidase, and enhanced chemiluminescence (Amersham). Anti-panβ and anti-Gβ5 rabbit polyclonal antibodies were obtained from Chemicon; anti-Gγ2 rabbit antiserum was purchased from Santa Cruz Biotechnology.
GAP Assays.
G protein subunits used as substrates for GAP assays were expressed in Escherichia coli or Sf9 cells and purified as described (8, 10). Single turnover GTPase assays were conducted as described (11–13); the concentrations of substrates are listed in the legend to Fig. 6.
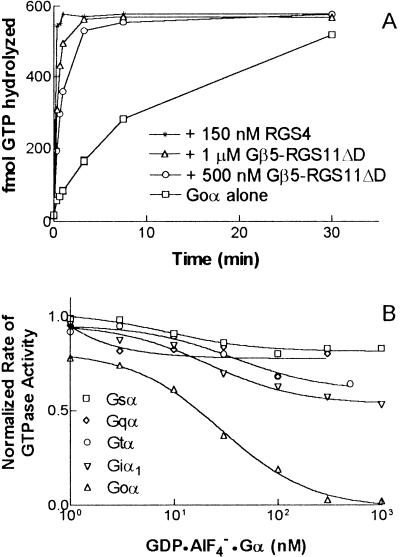
Figure 6.
The Gβ5/RGS11ΔD heterodimer is a GAP for Goα. (A) Myristoylated Goα bound with [γ-32P]GTP (90 nM) served as the substrate for Gβ5/RGS11ΔD in single turnover GAP assays conducted in solution at 4°C. Production of 32Pi was monitored after the addition of Mg2+ to initiate the reaction and either 0, 500 nM, or 1 μM Gβ5/RGS11ΔD. Reactions containing 150 nM RGS4 served as positive controls. Data shown are representative of more than three separate experiments. (B) Inhibition of the Gβ5/RGS11ΔD-stimulated GTPase activity of Goα by transition-state complexes of various Gα subunits. Transition-state (GDP-AlF4−) complexes of myristoylated Goα, myristoylated Giα1, Gsα, Gqα, and Gtα were incubated with Gβ5/RGS11ΔD for 30 min on ice in a buffer containing 10 mM NaF, 5 mM MgCl2, and 20 μM AlCl3. This mixture was then diluted 10-fold by addition of [γ-32P]GTP-Goα in buffer containing 40 μM GTP, 5.5 mM 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate, 50 mM NaHepes (pH 8.0), 1 mM DTT, 1 mM EDTA, 0.1 mg/ml of BSA, and 4% glycerol. The final concentrations of myristoylated Goα-GTP substrate and Gβ5/RGS11ΔD were 200 nM; the final concentrations of the competing Gα-transition state complexes are indicated. Each point represents the initial rate of GTP hydrolysis, determined by fitting a nine-point time course to a linear regression. The initial rate of GTP hydrolysis by Goα was 0.04/min in the absence of Gβ5/RGS11ΔD and 0.2/min in its presence.
RESULTS
RGS11 Shares a GGL Sequence With Other RGS Proteins.
Koelle and Horvitz (14) first described an ≈200 bp span of rat RGS11. We identified sequence with 87% identity to rat RGS11 within human genomic cosmid DNA derived from chromosome 16p13.3 (EMBL accession no. Z69667). By removal of introns, this genomic sequence was used to predict the RGS11 cDNA sequence encoding the RGS domain. To determine the 5′-end of RGS11 cDNA, rapid amplification of cDNA ends reactions were performed on human brain cDNA as described (15). The 3′-end was identified by blast searches of human expressed sequence tag databases by using cosmid sequence 3′ of the RGS domain (EMBL accession no. Z69667). Several expressed sequence tags were identical to cosmid sequence and contained putative polyadenylation signals and poly(A) tails (e.g., EMBL accession no.Z39463 and GenBank accession no. AA907380). To verify that the 5′ rapid amplification of cDNA ends products and 3′ expressed sequence tag sequences formed a contiguous mRNA, we amplified the entire RGS11 cDNA using the reverse transcription–PCR on human retinal RNA (7). The RGS11 cDNA (GenBank accession no. AF035153) encodes an ORF of 467 aa with a predicted molecular weight of 53,000. The entire RGS11 gene is contained within known cosmid sequences from human chromosome 16p13.3 (EMBL accession no. Z69667 and GenBank accession no. AC004754).
The amino acid sequence of RGS11 is most similar to that of RGS9 (16, 17). The RGS11 amino terminus (aa 32–132) encodes a DEP domain (Dishevelled, EGL-10, Pleckstrin) of unknown function (18) that is 83% similar to the DEP domain of murine RGS9, whereas the RGS domain of RGS11 (aa 299–414) is 77% similar to that of RGS9. blast searches with sequences outside the DEP or RGS domains revealed a 64 aa region (aa 219–282) that is not only conserved in location and sequence with similar regions of RGS6, RGS7, RGS9, and EGL-10, but is also 34% identical to the G protein γ5 subunit. Inclusion of other G protein γ subunits in subsequent multiple sequence alignments led us to designate this region of RGS11 as the GGL domain (Fig. 1).
Figure 1.
Sequence alignment between Gγ subunits and the GGL domains of RGS6, RGS7, RGS9, RGS11, and EGL-10. Residues conserved among RGS proteins are in black boxes; residues in common between RGS proteins and one or both Gγ consensus lines (Cons-A, Cons-B) are shown in shaded boxes. Asterisk denotes first residue of the RGS domain. b, bovine; c, C. elegans; h, human; m, mouse; r, rat.
The GGL Domain Binds to G Protein β5 Subunits.
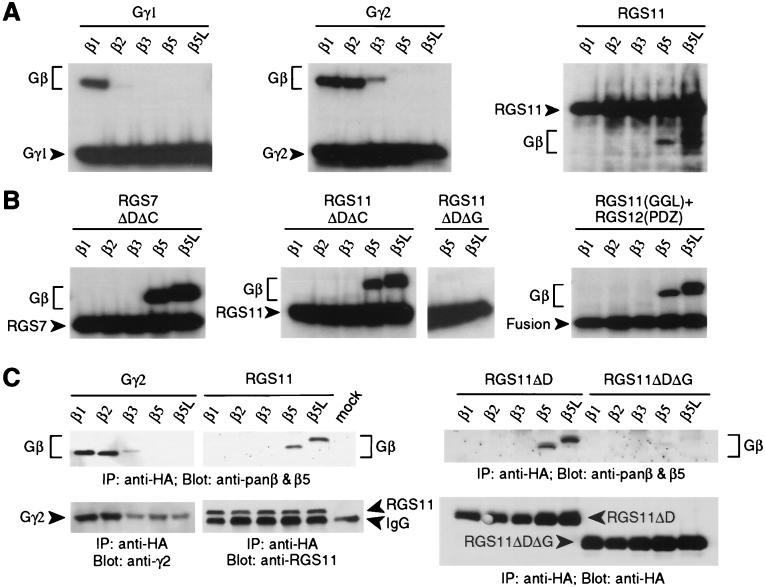
N-terminal extension of Gγ subunits with the influenza virus HA epitope does not affect formation of either βγ dimers or G protein heterotrimers (19). Hence, we produced N-terminally HA-tagged Gγ or GGL domain-containing RGS proteins by in vitro transcription/translation in combination with various Gβ subunits to detect possible interactions. 35S-labeled Gγ and RGS proteins were immunoprecipitated by using an anti-HA mAb. Associated, 35S-labeled Gβ subunits were detected by SDS/PAGE and autoradiography. Gγ1 bound solely to Gβ1, whereas Gγ2 bound to both Gβ1 and Gβ2 (Fig. 2A), as described (20, 21). Weak interaction was also detected between Gγ2 and Gβ3 (Fig. 2A and below). In contrast, RGS11 did not interact with Gβ1, Gβ2, or Gβ3; however, both Gβ5 and the longer, retinal-specific isoform Gβ5L (22) were both coimmunoprecipitated with RGS11 (Fig. 2A). Similar results were obtained with RGS7 and RGS11 proteins truncated to contain only the GGL and RGS domains (ΔDΔC; Fig. 2B). Association of RGS11 with Gβ5 and Gβ5L is dependent on the GGL domain, because no Gβ subunits were coimmunoprecipitated with a truncated RGS11 polypeptide containing only the RGS domain (RGS11ΔDΔG; Fig. 2B).
Figure 2.
Gβ binding specificity of the GGL domain. HA-tagged Gγ or RGS proteins were either cotranslated in vitro (A and B) or cotransfected into COS-7 cells (C) with individual Gβ subunits, immunoprecipitated (IP) with anti-HA mAb, and visualized by SDS/PAGE and autoradiography (A and B) or immunoblotting (C) with indicated antisera (Blot). (A) Gβ subunit association in vitro with Gγ1, Gγ2, and full-length RGS11 proteins. (B) Gβ subunit association in vitro with truncated RGS7 protein (ΔDΔC, aa 202–395 of SwissProt accession no.P49802), truncated RGS11 proteins (ΔDΔC, aa 219–423; ΔDΔG, aa 283–467), and a chimeric protein (Fusion) composed of the RGS11 GGL domain (aa 219–283) fused to the rat RGS12 PDZ domain (aa 1–91 of SwissProt accession no. O08774). (C) Gβ subunit association with Gγ2, full-length RGS11, and truncated RGS11 proteins (ΔD, aa 219–467; ΔDΔG, aa 283–467) in COS-7 cells.
The GGL domain alone was poorly expressed in vitro and in cell transfection systems (data not shown). To ascertain whether the GGL sequence is an autonomous Gβ5-binding domain, we tested fusions between the RGS11 GGL domain and the PDZ or RGS domains of RGS12 (4) for their ability to interact with Gβ subunits. Both Gβ5 and Gβ5L were coimmunoprecipitated with the GGL/PDZ and GGL/RGS fusion proteins (Fig. 2B and data not shown). This binding is not mediated by the RGS12-derived fusion partners; full-length RGS12 did not interact with Gβ subunits (data not shown).
To demonstrate binding of the GGL domain to Gβ5 subunits in a cellular context, COS-7 cells were transiently cotransfected with expression vectors encoding various Gβ subunits and either HA-tagged Gγ2 or RGS11. Cell lysates were immunoprecipitated with anti-HA mAb, and associated Gβ subunits were detected by immunoblotting using a mixture of pan-Gβ and Gβ5-specific polyclonal antisera. Gγ2 associated with Gβ1, Gβ2, and weakly with Gβ3, but not with Gβ5 or Gβ5L (Fig. 2C). In contrast, only Gβ5 and Gβ5L were coimmunoprecipitated with full-length RGS11 and truncated RGS11 lacking the DEP domain (RGS11ΔD; Fig. 2C). Further truncation of RGS11, deleting the GGL domain, abolished detectable binding to Gβ5 and Gβ5L (RGS11ΔDΔG; Fig. 2C).
Expression of RGS11 and Gβ5 in Sf9 Cells.
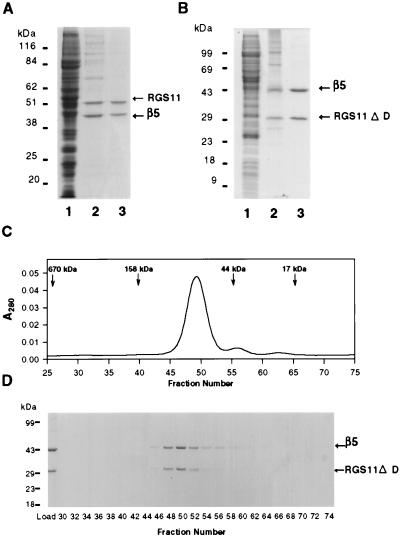
Recombinant baculoviruses encoding either RGS11 or RGS11ΔD and either hexahistidine-tagged Gβ5 or Gβ5L were used to direct protein expression in Sf9 cells. Immunoblotting of Sf9 cell fractions indicated accumulation of both RGS11 and Gβ5 in both particulate and cytoplasmic fractions (not shown). Chromatography of the cytoplasmic fraction over a column of Ni-NTA agarose resulted in coincidental enrichment of both the hexahistidine-tagged Gβ5 subunit and untagged RGS11 (Fig. 3A) or RGS11ΔD (Fig. 3B). The two proteins also comigrated during chromatography over a Pharmacia Mono Q HR5/5 column (Fig. 3 A and B) and during gel filtration (Fig. 3 C and D for Gβ5 and RGS11ΔD). Gel filtration of Gβ5/RGS11 demonstrated that the complex of the full-length proteins was aggregated. The complex between RGS11 and Gβ5 is clearly a stable one, and quantities of the purified heterodimeric complex sufficient for biochemical analysis can be obtained by expression in Sf9 cells.
Figure 3.
Purification of Gβ5/RGS11 heterodimers after expression in Sf9 cells. Cells were infected with recombinant baculoviruses encoding either (A) hexahistidine-tagged Gβ5 and full-length RGS11 or (B) hexahistidine-tagged Gβ5 and RGS11ΔD (aa 219–467). Fractions were subjected to electrophoresis through polyacrylamide gels containing sodium dodecylsulfate and stained with Coomassie blue. Lanes: 1, 15 μg of soluble lysate; 2, 1 μg of Ni-NTA eluate; 3, 1 μg of Mono Q eluate. (C) The peak from the Mono Q column shown in B (150 μg of protein) was chromatographed over a Pharmacia 16/60 Superdex 200 gel filtration column, and fractions (0.2 ml) were monitored for absorbance at 280 nm or (D) were analyzed electrophoretically and stained with Coomassie blue.
Molecular Modeling of the GGL/Gβ5 Interface.
We constructed a model of the interface between the GGL domain of RGS11 and Gβ5, starting with the crystal structures of Gγ1 and Gβ1 (23, 24). Structural predictions are based on the assumption that the GGL domain binds to the hydrophobic cleft of Gβ5 in a manner analogous to the interaction between Gβ1 and either Gγ1 or Gγ2. GGL and Gβ5 side chains that differed from those of Gγ1 and Gβ1, respectively, were modeled by using the program o (25). The model of the heterodimer was then refined with 200 steps of positional refinement, followed by a molecular dynamics simulation (500 steps at 300 K for a total of 0.4 psec) by using the program cns (26). The amino acid sequences of Gβ5 and the GGL domain of RGS11 appear well suited for their high-affinity interaction (Fig. 4). We believe that the GGL domain will not interact with Gβ2 or Gβ3 because of Tyr-251 in RGS11. A Phe residue in this position of Gγ1 prevents functional pairing with either Gβ2 or Gβ3 (21). Other residues unique to Gβ5 that may be important for the selectivity of GGL for Gβ5 include Val-274 (Leu-261 in Gβ1–4) and Ala-353 (Asn-340 in Gβ1–4). Val-274 and Ala-353 of Gβ5 are smaller than their counterparts in Gβ1–4 and avoid collisions with the side chains of Leu-248 and Trp-274, respectively from RGS11. Analogous residues in Gγ subunits are Gly, Ala, Cys, or Ser instead of Leu-248, and Phe instead of Trp-274.
Figure 4.
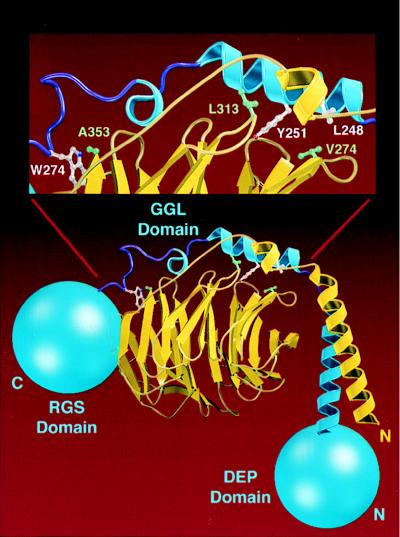
Molecular modeling of the Gβ5/GGL domain interface. The model of Gβ5 (yellow) associated with the GGL domain of RGS11 (blue) is shown with the N-terminal DEP and C-terminal RGS domains of RGS11 drawn as blue spheres. (Inset) Specific residues at the Gβ5–GGL interface. Residues Val-274, Leu-313, and Ala-353 from Gβ5, which are thought to be important for specificity, are green. Analogous residues Leu-248, Tyr-251, and Trp-274 from RGS11 are white.
mRNA for RGS11 and Gβ5 Have a Similar Tissue Distribution.
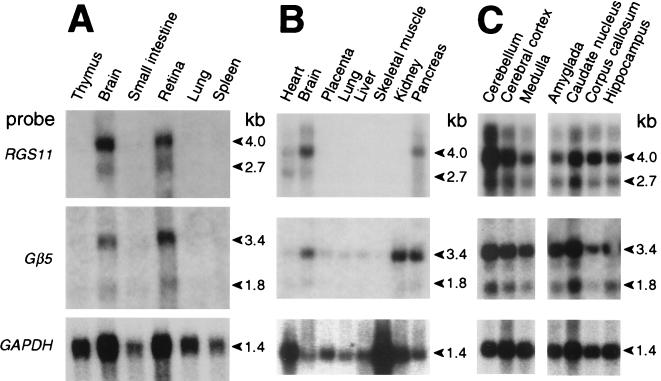
In contrast to the nearly ubiquitous expression of other Gβ subunits, Gβ5 isoforms are expressed in a highly tissue-restricted fashion; mouse Gβ5 is expressed predominantly in the brain, with transcripts also detectable in the kidney (27), whereas mouse Gβ5L is largely confined to the retina (22). We have performed Northern blot analysis on RNA isolated from human tissues. RGS11 and Gβ5 mRNA were expressed in overlapping patterns, with high levels of transcripts seen in both brain and retina (Fig. 5A) and lower amounts in the pancreas (Fig. 5B). Messenger RNA for Gβ5, but not RGS11, was detected in the kidney, whereas the reverse was observed in the heart (Fig. 5B). Both RGS11 and Gβ5 transcripts were seen in all brain anatomical regions tested, with highest relative expression of RGS11 in the cerebellum (Fig. 5C and data not shown). This widespread expression of RGS11 mRNA in the human brain is in contrast to the restricted pattern observed by Gold and colleagues in the rat brain (28).
Figure 5.
Northern blot analyses of RGS11 and Gβ5 expression patterns. Blots of (A) 20 μg total RNA or (B and C) 2 μg poly(A+) RNA from various human tissues were serially hybridized with a human RGS11 cDNA probe, a mouse Gβ5 cDNA probe, and, as a control for RNA loading and quality, a human glyceraldehyde-3-phosphate dehydrogenase probe.
Functions of the Gβ5/RGS11 Heterodimer.
The existence of a complex containing RGS and GGL domains as well as a Gβ subunit suggests many possible functions, including GAP activity toward G protein α subunits, interactions with Gα proteins by means of the Gβ5 subunit, and the many varied activities of Gβγ complexes themselves. To date, the only such activity detected is the capacity of the Gβ5/RGS11 complex to exert GAP activity selectively toward the GTP–Goα complex. The capacity of the complex to exert GAP activity on GTP–Gα substrates was examined in single turnover assays in solution. Enhanced GTPase activity of Goα was detected with either Gβ5/RGS11 (not shown) or with concentrations of Gβ5/RGS11ΔD as low as 10 nM; addition of 1 μM concentrations of the complex increased the single turnover rate for GTP hydrolysis from 0.13 min−1 to 3.2 min−1 at 4°C (Fig. 6A). The capacity of the complex to accelerate the GTPase activity of Giα1–3 was very modest (2-fold; not shown), and GAP activity was not detected with Gsα, Gqα, Gzα, G12α, or G13α as substrates (not shown). The capacity of GDP-AlF4−-bound G protein α subunits to inhibit the GAP activity of Gβ5/RGS11ΔD toward Goα was also examined. This is a measure of affinity of the transition state-like complex of the α subunit for the RGS protein in question (12, 29). These assays indicated an apparent affinity of GDP-AlF4−-Goα for Gβ5/RGS11ΔD of roughly 10–100 nM and no detectable affinity of the transition state (GDP-AlF4−) complexes of Gsα, Giα1, Gqα, or Gtα for Gβ5/RGS11ΔD (Fig. 6B).
DISCUSSION
In almost all cases examined to date the existence of an RGS domain in a protein has adequately predicted GAP activity of that protein toward a heterotrimeric G protein α subunit of the Gi, Gq, and/or G12 subfamily. It is now clear that the story is not that simple. At least some RGS proteins have repertoires that are more complex than simple negative regulation of G protein-mediated signaling pathways. For example, the RGS domain in the rho guanine nucleotide exchange factor p115 imparts sensitivity (to p115) to regulation of its activity by Gα13 and Gα12 (5, 6). RGS proteins can thus serve as effectors for G protein action. We have now described the existence in RGS11 and RGS7 (and possibly in RGS6 and RGS9 and EGL-10) of a GGL domain that imparts to these proteins the capacity to form heterodimers with the G protein β5 subunit. This complex appears to have unusually selective GAP activity toward Goα, although the specificity of the GAP activity of RGS proteins determined in solution (in vitro) has not always proven reliable (13).
The assembly of this complex, containing DEP and RGS domains in addition to its Gβγ-like core, suggests functional consequences. Most obvious are the interactions of typical Gβγ heterodimers with GDP-bound Gα proteins; a variety of effectors, including adenylyl cyclases, phospholipases, and ion channels; and regulatory proteins such as phosducin and receptor kinases. We have failed to date to detect interactions of Gβ5/RGS11 with GDP-bound G protein α subunits, adenylyl cyclases, and phospholipases. Such interactions may not be characteristic of this atypical Gβγ-like complex, perhaps because of interference by the appended RGS and DEP domains. For example, the model shown in Fig. 4 suggests that the RGS domain of RGS11 could lie near the binding site on Gβ for the amino terminus of Gα proteins (23, 24). Perhaps then such interactions might be evident only after hypothetical regulatory modifications of the Gβ5/RGS11 heterodimer. If Gβ5/RGS11 and other related complexes (e.g., between Gβ5 and RGS6, RGS7, and RGS9) do participate in interactions that typify other G protein βγ heterodimers, the inclusion of the RGS domain in these complexes will presumably have functional consequences for the signaling pathways involved, and the significance of these complexes may lie in the assembly of molecular machines that can perform signaling reactions with appropriate kinetic properties and specificity.
We must also consider the possibility that the Gβ5 subunit is atypical and that its general functions may not be deduced by comparison with the other four Gβ proteins. Gβ1–4 are very similar structurally (80–90% sequence identity); Gβ5 is a clear outlier—only about 50% identical to Gβ1–4. Gβ1–4 are membrane-bound proteins; Gβ5 in retina is soluble (although Gβ5L in retina is particulate), and a fraction of Gβ5 in brain may also be soluble (22). Nevertheless, coexpression of Gβ5 with conventional Gγ subunits does cause typical Gβγ-like effects, such as activation of phospholipase C-β (27), and Gβ5 appears to interact reasonably well with Gγ3 and 4 in yeast two hybrid assays (30). Although Fletcher et al. (31) observed selective interactions of Gαq with a complex of Gβ5 and Gγ2, they also noted that this βγ complex dissociated at concentrations of sodium cholate in excess of 0.05%—atypical behavior for a βγ complex. There is a clear need to learn the identity of the partners of both Gβ5 and RGS6, RGS7, RGS9, and RGS11 in vivo. Gβ5 may associate with conventional γ subunits, GGL-containing RGS proteins, and other targets for β subunits, whereas the GGL-containing RGS proteins may enjoy a wealth of molecular interactions by means of their GGL, RGS, and DEP domains.
Acknowledgments
We thank Denis Bouchard for technical assistance, Bethany Sutton for sequencing support, Mike Bass for computational support, and Bryan Sutton and Axel Brunger for assistance with molecular modeling. This work was supported in part by Amgen (D.P.S.), National Institutes of Health Grant GM34497 (A.G.G.), and the Raymond and Ellen Willie Distinguished Chair in Molecular Neuropharmacology (A.G.G.).
ABBREVIATIONS
- DEP
Dishevelled/Egl-10/Pleckstrin
- GGL
G-gamma-like
- RGS
regulator of G-protein signaling
- ΔD
DEP domain deletion
- ΔC
C-terminus deletion
- ΔG
GGL domain deletion
- GAPs
GTPase-activating proteins
- HA
hemagglutinin
- PDZ
PSD-95/Dlg/Z0-1
Footnotes
References
- 1. Dohlman H G, Thorner J. J Biol Chem. 1997;272:3871–3874. doi: 10.1074/jbc.272.7.3871. [DOI] [PubMed] [Google Scholar]
- 2.Koelle M R. Curr Opin Cell Biol. 1997;9:143–147. doi: 10.1016/s0955-0674(97)80055-5. [DOI] [PubMed] [Google Scholar]
- 3.Berman D M, Gilman A G. J Biol Chem. 1998;273:1269–1272. doi: 10.1074/jbc.273.3.1269. [DOI] [PubMed] [Google Scholar]
- 4.Snow B E, Hall R A, Krumins A M, Brothers G M, Bouchard D, Brothers C A, Chung S, Mangion J, Gilman A G, Lefkowitz R J, Siderovski D P. J Biol Chem. 1998;273:17749–17755. doi: 10.1074/jbc.273.28.17749. [DOI] [PubMed] [Google Scholar]
- 5.Kozasa T, Jiang X, Hart M J, Sternweis P M, Singer W D, Gilman A G, Bollag G, Sternweis P C. Science. 1998;280:2109–2111. doi: 10.1126/science.280.5372.2109. [DOI] [PubMed] [Google Scholar]
- 6.Hart M J, Jiang X, Kozasa T, Roscoe W, Singer W D, Gilman A G, Sternweis P C, Bollag G. Science. 1998;280:2112–2114. doi: 10.1126/science.280.5372.2112. [DOI] [PubMed] [Google Scholar]
- 7.Snow B E, Antonio L, Suggs S, Siderovski D P. Gene. 1998;206:247–253. doi: 10.1016/s0378-1119(97)00593-3. [DOI] [PubMed] [Google Scholar]
- 8.Lee E, Linder M E, Gilman A G. Methods Enzymol. 1994;237:146–164. doi: 10.1016/s0076-6879(94)37059-1. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt C J, Neer E J. J Biol Chem. 1991;266:4538–4544. [PubMed] [Google Scholar]
- 10.Kozasa T, Gilman A G. J Biol Chem. 1995;270:1734–1741. doi: 10.1074/jbc.270.4.1734. [DOI] [PubMed] [Google Scholar]
- 11.Berman D M, Wilkie T M, Gilman A G. Cell. 1996;86:445–452. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- 12.Berman D M, Kozasa T, Gilman A G. J Biol Chem. 1996;271:27209–27212. doi: 10.1074/jbc.271.44.27209. [DOI] [PubMed] [Google Scholar]
- 13.Ingi T, Krumins A, Chidiac P, Brothers G M, Chung S, Snow B E, Barnes C A, Lanahan A A, Siderovski D P, Ross E M, Gilman A G, Worley P F. J Neurosci. 1998;18:7178–7188. doi: 10.1523/JNEUROSCI.18-18-07178.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koelle M R, Horvitz H R. Cell. 1996;84:115–125. doi: 10.1016/s0092-8674(00)80998-8. [DOI] [PubMed] [Google Scholar]
- 15.Snow B E, Antonio L, Suggs S, Gutstein H B, Siderovski D P. Biochem Biophys Res Commun. 1997;233:770–777. doi: 10.1006/bbrc.1997.6537. [DOI] [PubMed] [Google Scholar]
- 16.He W, Cowan C W, Wensel T G. Neuron. 1998;20:95–102. doi: 10.1016/s0896-6273(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 17.Thomas E A, Danielson P E, Sutcliffe J G. J Neurosci Res. 1998;52:118–124. doi: 10.1002/(SICI)1097-4547(19980401)52:1<118::AID-JNR11>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 18.Ponting C P, Bork P. Trends Biochem Sci. 1996;21:245–246. [PubMed] [Google Scholar]
- 19.Mende U, Schmidt C J, Spring D J, Neer E J. J Biol Chem. 1995;270:15892–15898. doi: 10.1074/jbc.270.26.15892. [DOI] [PubMed] [Google Scholar]
- 20.Pronin A N, Gautam N. Proc Natl Acad Sci USA. 1992;89:6220–6224. doi: 10.1073/pnas.89.13.6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt C J, Thomas T C, Levine M A, Neer E J. J Biol Chem. 1992;267:13807–13810. [PubMed] [Google Scholar]
- 22.Watson A J, Aragay A M, Slepak V Z, Simon M I. J Biol Chem. 1996;271:28154–28160. doi: 10.1074/jbc.271.45.28154. [DOI] [PubMed] [Google Scholar]
- 23.Wall M A, Coleman D E, Lee E, Iniguez-Lluhi J A, Posner B A, Gilman A G, Sprang S R. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 24.Lambright D G, Sondek J, Bohm A, Skiba N P, Hamm H E, Sigler P B. Nature (London) 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 25.Jones T A, Kjeldgard M. o. Upsala, Sweden: Uppsala Univ.; 1996. , Version 5.1. [Google Scholar]
- 26.Adams P D, Pannu N S, Read R J, Brunger A T. Proc Natl Acad Sci USA. 1997;94:5018–5023. doi: 10.1073/pnas.94.10.5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watson A J, Katz A, Simon M I. J Biol Chem. 1994;269:22150–22156. [PubMed] [Google Scholar]
- 28.Gold S J, Ni Y G, Dohlman H G, Nestler E J. J Neurosci. 1997;17:8024–8037. doi: 10.1523/JNEUROSCI.17-20-08024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tesmer J J G, Berman D M, Gilman A G, Sprang S R. Cell. 1997;89:251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- 30.Yan K, Kalyanaraman V, Gautam N. J Biol Chem. 1996;271:7141–7146. doi: 10.1074/jbc.271.12.7141. [DOI] [PubMed] [Google Scholar]
- 31.Fletcher J E, Lindorfer M A, Defilippo J M, Yasuda H, Guilmard M, Garrison J C. J Biol Chem. 1998;273:636–644. doi: 10.1074/jbc.273.1.636. [DOI] [PubMed] [Google Scholar]