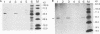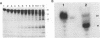Abstract
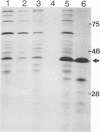
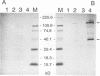
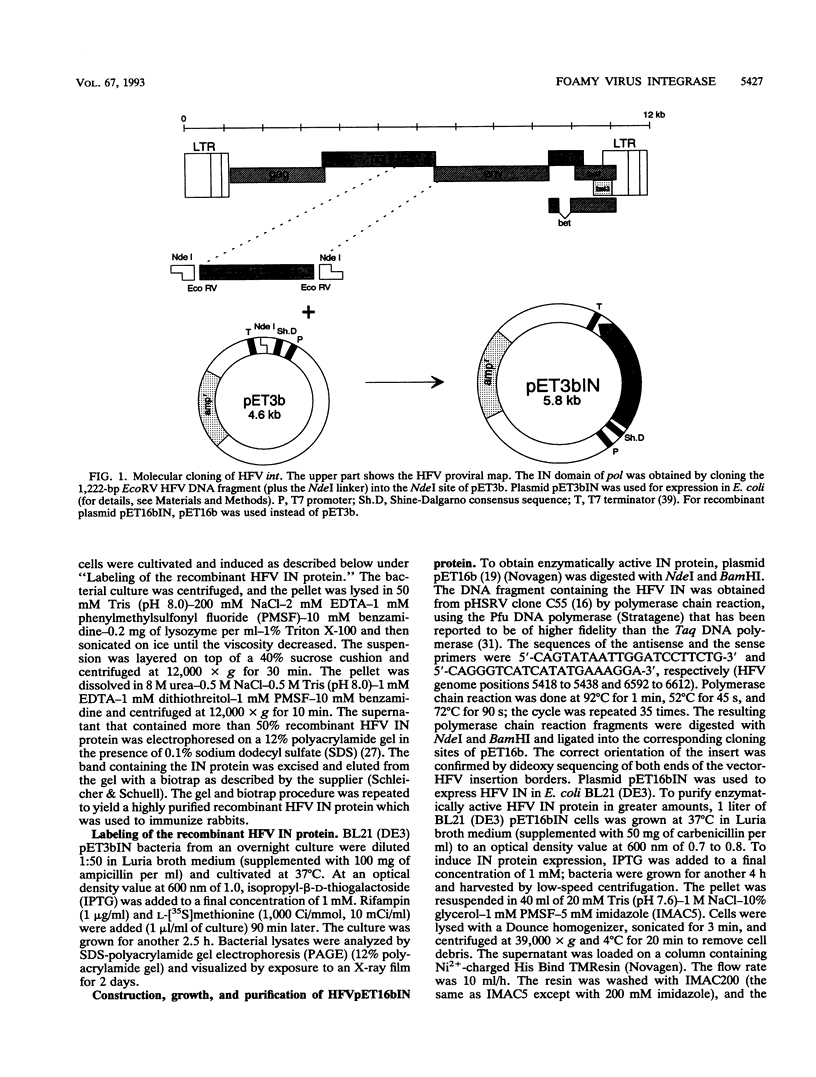
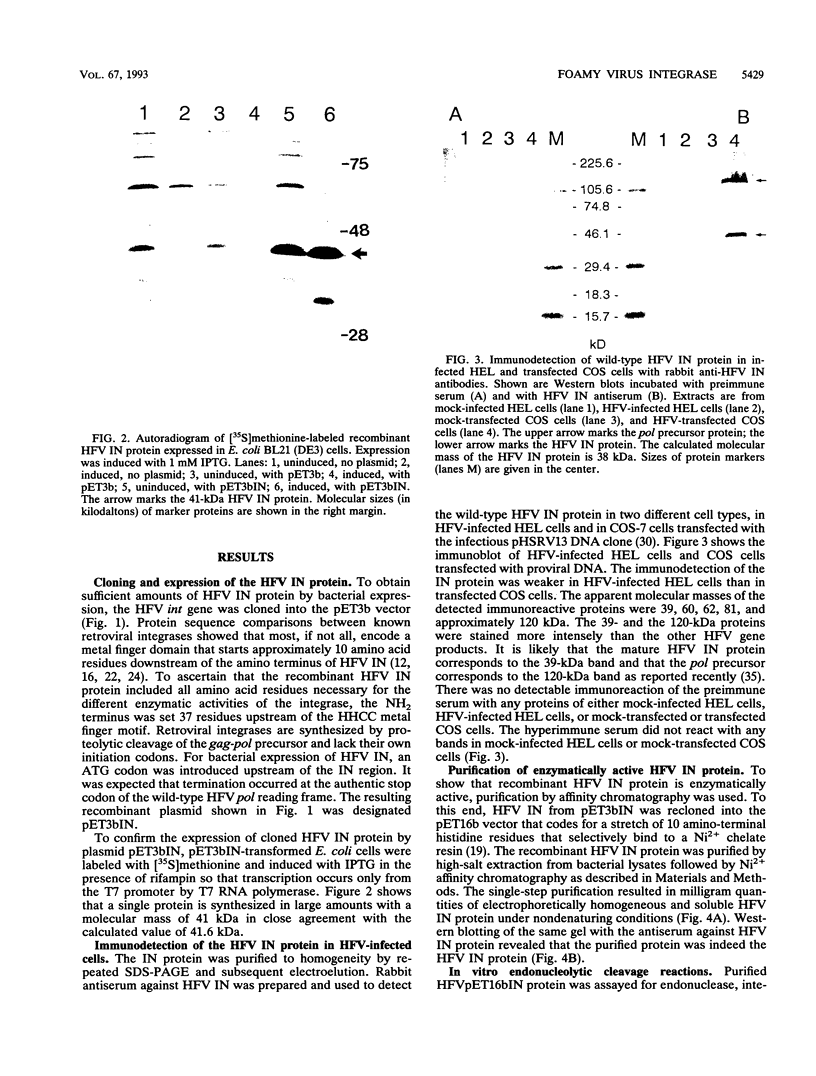
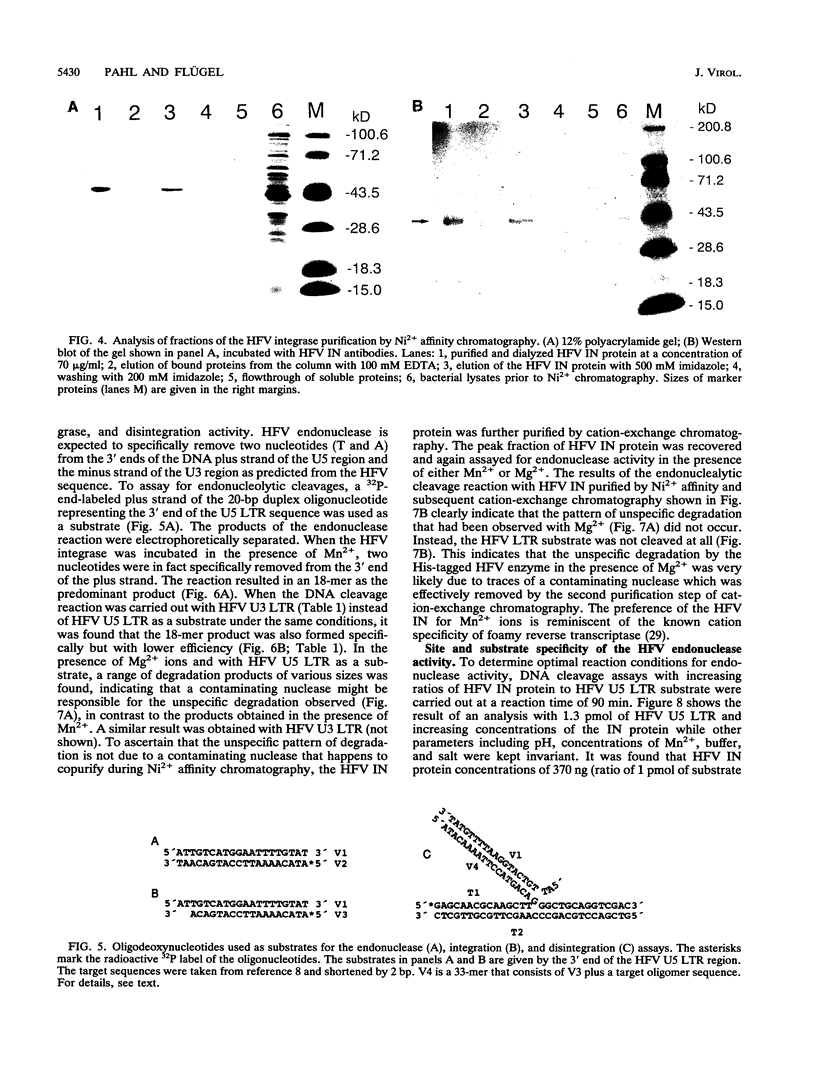
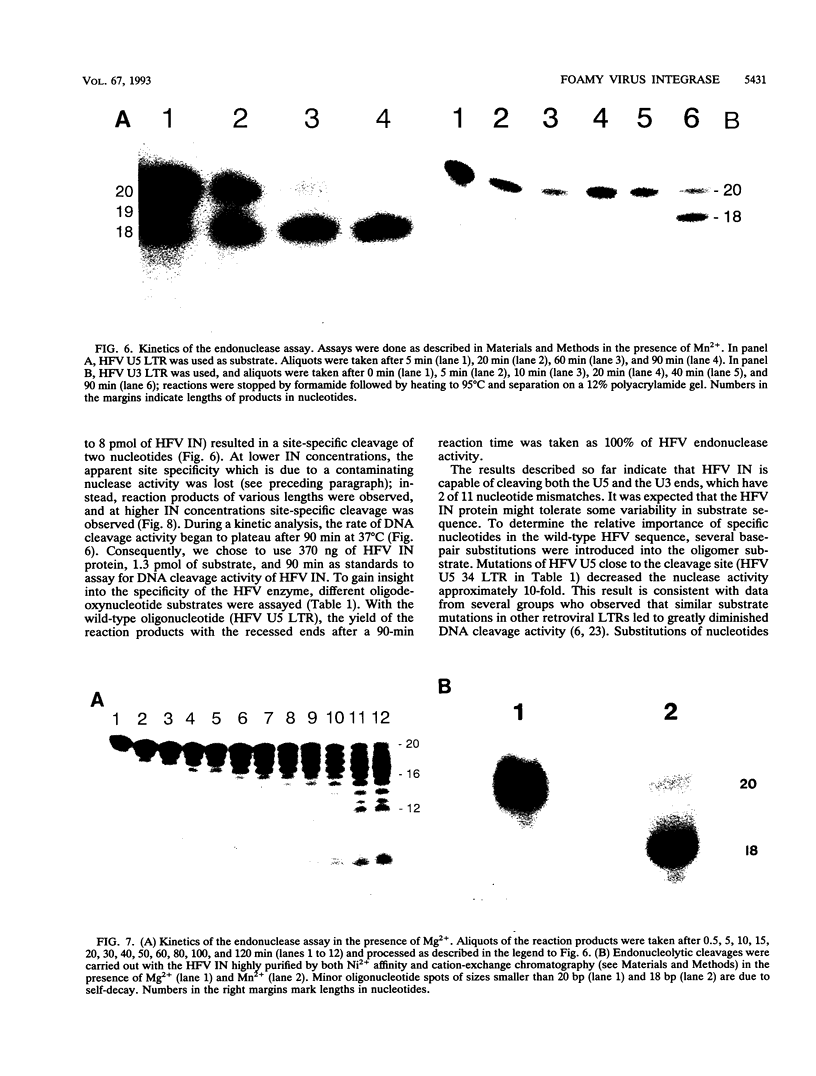
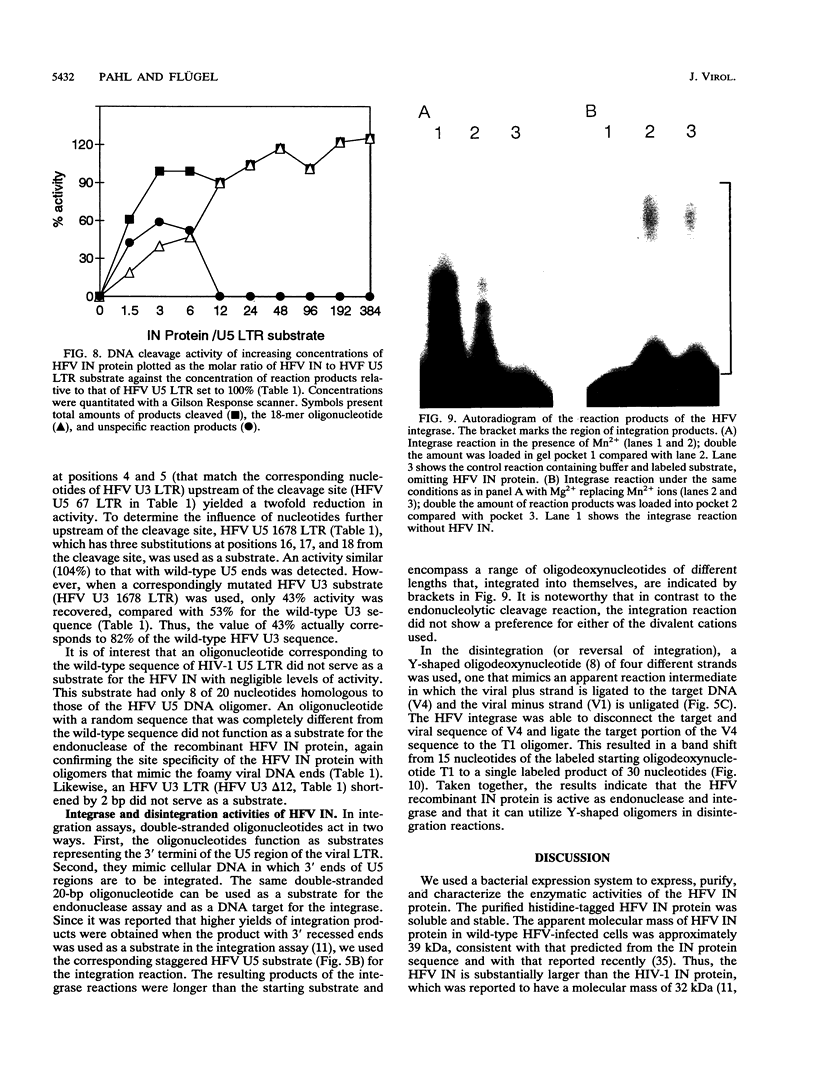
The bacterial expression plasmids, pET3b and pET16b, that contain the integrase domain of the human foamy virus (HFV) reverse transcriptase were constructed and expressed in Escherichia coli. The histidine-tagged HFV IN protein was purified to near homogeneity by single-step Ni2+ chelate affinity chromatography. HFV-specific proteins of 39 and 120 kDa from virus-infected cells reacted with antisera raised against the recombinant IN protein. Purified recombinant HFV IN protein was active as an endonuclease specifically cleaving two nucleotides from a 20-bp oligodeoxynucleotide substrate that mimics the authentic 5' ends of HFV DNA. Substrates with mutations relatively close to the cleavage site were less efficiently cleaved or not cleaved at all compared with the HFV U5 DNA end. The purified recombinant protein was active as integrase with double-stranded oligodeoxynucleotide substrates. The reverse reaction of DNA strand transfer, the disintegration activity, was shown by efficient cleavage of an intermediate Y-shaped oligodeoxynucleotide. In the presence of Mn2+ as the preferred divalent cation, oligodeoxynucleotides were specifically and efficiently cleaved. In contrast, endonucleolytic cleavages in the presence of Mg2+ ions led to a broad range of reaction products with the His-tagged HFV IN protein. After further purification of the HFV IN by cation-exchange chromatography, the unspecific degradation of oligonucleotide substrate in the presence of Mg2+ was not detectable.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achong B. G., Mansell P. W., Epstein M. A., Clifford P. An unusual virus in cultures from a human nasopharyngeal carcinoma. J Natl Cancer Inst. 1971 Feb;46(2):299–307. [PubMed] [Google Scholar]
- Bowerman B., Brown P. O., Bishop J. M., Varmus H. E. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989 Apr;3(4):469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brown P. O. Integration of retroviral DNA. Curr Top Microbiol Immunol. 1990;157:19–48. doi: 10.1007/978-3-642-75218-6_2. [DOI] [PubMed] [Google Scholar]
- Burke C. J., Sanyal G., Bruner M. W., Ryan J. A., LaFemina R. L., Robbins H. L., Zeft A. S., Middaugh C. R., Cordingley M. G. Structural implications of spectroscopic characterization of a putative zinc finger peptide from HIV-1 integrase. J Biol Chem. 1992 May 15;267(14):9639–9644. [PubMed] [Google Scholar]
- Bushman F. D., Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F. D., Fujiwara T., Craigie R. Retroviral DNA integration directed by HIV integration protein in vitro. Science. 1990 Sep 28;249(4976):1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- Chow S. A., Vincent K. A., Ellison V., Brown P. O. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science. 1992 Feb 7;255(5045):723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- Drelich M., Wilhelm R., Mous J. Identification of amino acid residues critical for endonuclease and integration activities of HIV-1 IN protein in vitro. Virology. 1992 Jun;188(2):459–468. doi: 10.1016/0042-6822(92)90499-f. [DOI] [PubMed] [Google Scholar]
- Engelman A., Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992 Nov;66(11):6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A., Mizuuchi K., Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991 Dec 20;67(6):1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- Flügel R. M., Rethwilm A., Maurer B., Darai G. Nucleotide sequence analysis of the env gene and its flanking regions of the human spumaretrovirus reveals two novel genes. EMBO J. 1987 Jul;6(7):2077–2084. doi: 10.1002/j.1460-2075.1987.tb02473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flügel R. M. Spumaviruses: a group of complex retroviruses. J Acquir Immune Defic Syndr. 1991;4(8):739–750. [PubMed] [Google Scholar]
- Fujiwara T., Craigie R. Integration of mini-retroviral DNA: a cell-free reaction for biochemical analysis of retroviral integration. Proc Natl Acad Sci U S A. 1989 May;86(9):3065–3069. doi: 10.1073/pnas.86.9.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandgenett D. P., Mumm S. R. Unraveling retrovirus integration. Cell. 1990 Jan 12;60(1):3–4. doi: 10.1016/0092-8674(90)90707-l. [DOI] [PubMed] [Google Scholar]
- Hoffmann A., Roeder R. G. Purification of his-tagged proteins in non-denaturing conditions suggests a convenient method for protein interaction studies. Nucleic Acids Res. 1991 Nov 25;19(22):6337–6338. doi: 10.1093/nar/19.22.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. S., McClure M. A., Feng D. F., Gray J., Doolittle R. F. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson C. B., Donzella G. A., Roth M. J. Characterization of the forward and reverse integration reactions of the Moloney murine leukemia virus integrase protein purified from Escherichia coli. J Biol Chem. 1993 Jan 15;268(2):1462–1469. [PubMed] [Google Scholar]
- Katz R. A., Merkel G., Kulkosky J., Leis J., Skalka A. M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990 Oct 5;63(1):87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- Katzman M., Katz R. A., Skalka A. M., Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol. 1989 Dec;63(12):5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan E., Mack J. P., Katz R. A., Kulkosky J., Skalka A. M. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 1991 Feb 25;19(4):851–860. doi: 10.1093/nar/19.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkosky J., Jones K. S., Katz R. A., Mack J. P., Skalka A. M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992 May;12(5):2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupiec J. J., Kay A., Hayat M., Ravier R., Périès J., Galibert F. Sequence analysis of the simian foamy virus type 1 genome. Gene. 1991 May 30;101(2):185–194. doi: 10.1016/0378-1119(91)90410-d. [DOI] [PubMed] [Google Scholar]
- LaFemina R. L., Callahan P. L., Cordingley M. G. Substrate specificity of recombinant human immunodeficiency virus integrase protein. J Virol. 1991 Oct;65(10):5624–5630. doi: 10.1128/jvi.65.10.5624-5630.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lundberg K. S., Shoemaker D. D., Adams M. W., Short J. M., Sorge J. A., Mathur E. J. High-fidelity amplification using a thermostable DNA polymerase isolated from Pyrococcus furiosus. Gene. 1991 Dec 1;108(1):1–6. doi: 10.1016/0378-1119(91)90480-y. [DOI] [PubMed] [Google Scholar]
- Löchelt M., Zentgraf H., Flügel R. M. Construction of an infectious DNA clone of the full-length human spumaretrovirus genome and mutagenesis of the bel 1 gene. Virology. 1991 Sep;184(1):43–54. doi: 10.1016/0042-6822(91)90820-2. [DOI] [PubMed] [Google Scholar]
- Maurer B., Bannert H., Darai G., Flügel R. M. Analysis of the primary structure of the long terminal repeat and the gag and pol genes of the human spumaretrovirus. J Virol. 1988 May;62(5):1590–1597. doi: 10.1128/jvi.62.5.1590-1597.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergia A., Luciw P. A. Replication and regulation of primate foamy viruses. Virology. 1991 Oct;184(2):475–482. doi: 10.1016/0042-6822(91)90417-a. [DOI] [PubMed] [Google Scholar]
- Mergia A., Shaw K. E., Lackner J. E., Luciw P. A. Relationship of the env genes and the endonuclease domain of the pol genes of simian foamy virus type 1 and human foamy virus. J Virol. 1990 Jan;64(1):406–410. doi: 10.1128/jvi.64.1.406-410.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netzer K. O., Schliephake A., Maurer B., Watanabe R., Aguzzi A., Rethwilm A. Identification of pol-related gene products of human foamy virus. Virology. 1993 Jan;192(1):336–338. doi: 10.1006/viro.1993.1039. [DOI] [PubMed] [Google Scholar]
- Renne R., Friedl E., Schweizer M., Fleps U., Turek R., Neumann-Haefelin D. Genomic organization and expression of simian foamy virus type 3 (SFV-3). Virology. 1992 Feb;186(2):597–608. doi: 10.1016/0042-6822(92)90026-l. [DOI] [PubMed] [Google Scholar]
- Sherman P. A., Dickson M. L., Fyfe J. A. Human immunodeficiency virus type 1 integration protein: DNA sequence requirements for cleaving and joining reactions. J Virol. 1992 Jun;66(6):3593–3601. doi: 10.1128/jvi.66.6.3593-3601.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Vincent K. A., Ellison V., Chow S. A., Brown P. O. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J Virol. 1993 Jan;67(1):425–437. doi: 10.1128/jvi.67.1.425-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink C., van Gent D. C., Elgersma Y., Plasterk R. H. Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J Virol. 1991 Sep;65(9):4636–4644. doi: 10.1128/jvi.65.9.4636-4644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]