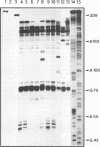
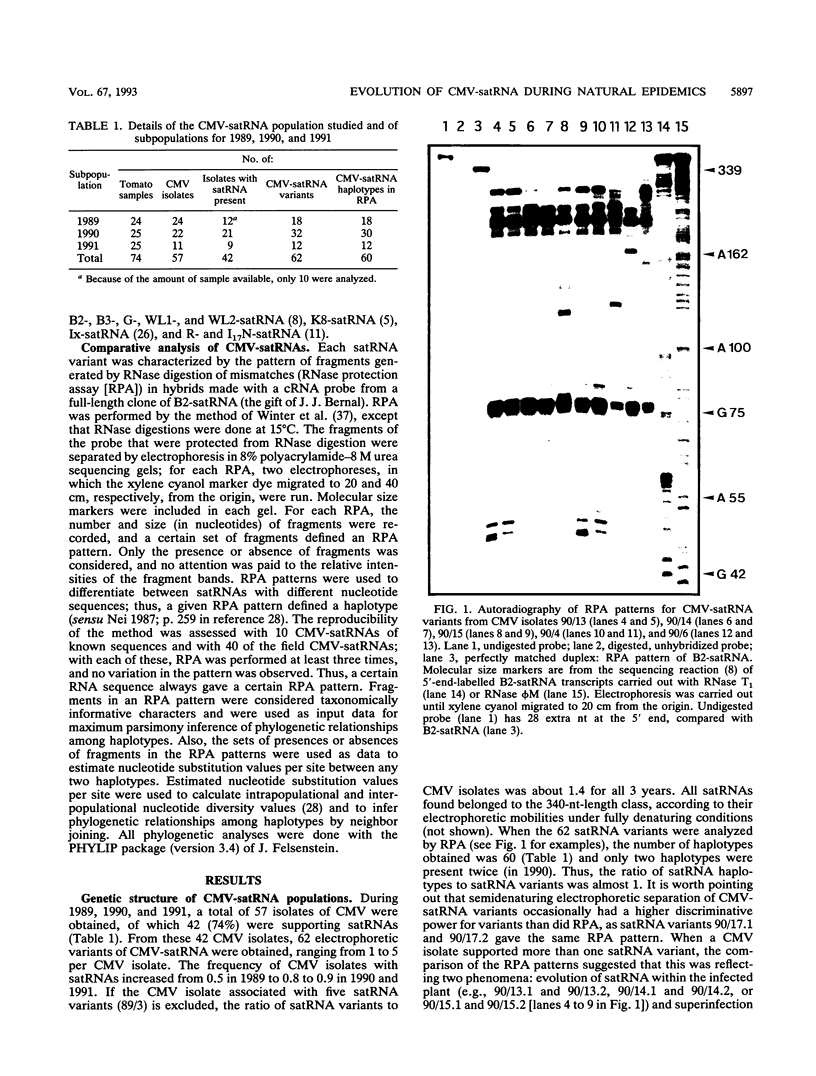
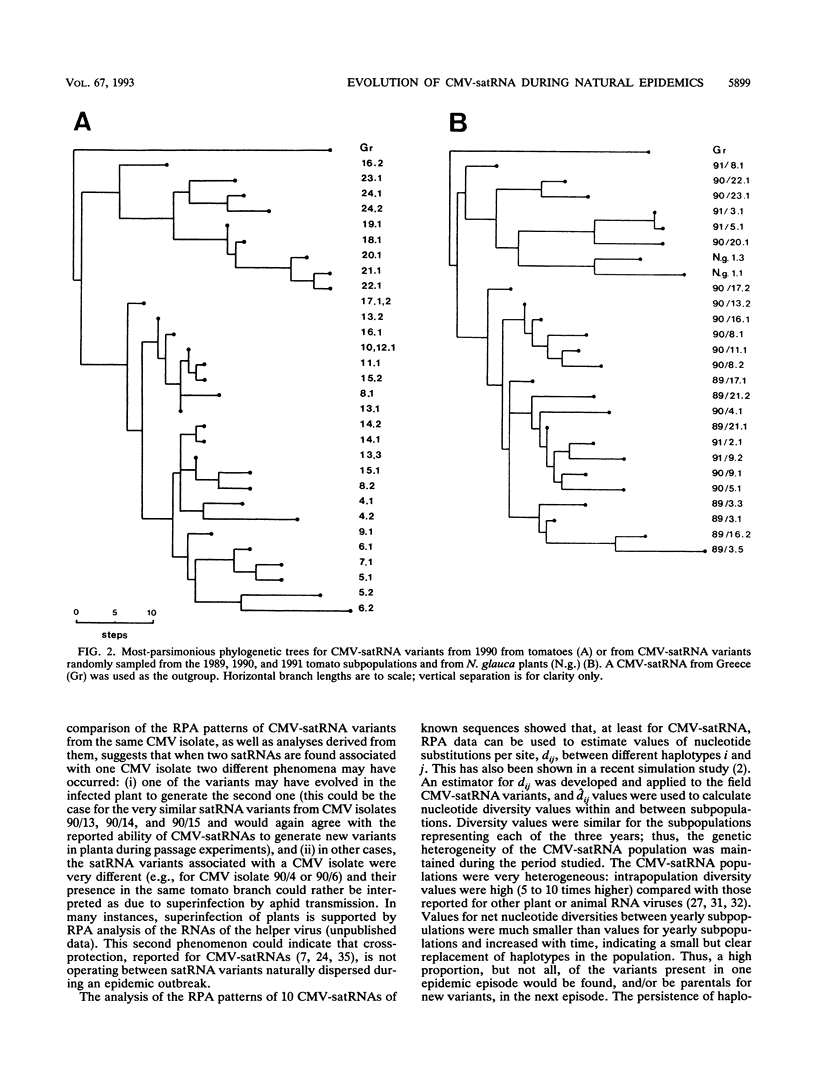
Abstract
The genetic structure of populations of cucumber mosaic virus (CMV) satellite RNA (satRNA) and its evolution were analyzed during the course of a CMV epidemic in tomatoes in eastern Spain. A total of 62 variants of CMV-satRNA from epidemic episodes in 1989, 1990, and 1991 were characterized by RNase protection assay (RPA); RPA patterns defined 60 haplotypes in the CMV-satRNA population. RPA of nine CMV-satRNAs of known sequences showed that numbers of nucleotide substitutions per site (dij) between different satRNAs can be estimated from RPA data. Thus, dij were estimated for any possible pair of field CMV-satRNA types, and nucleotide diversities within and between yearly subpopulations were calculated. Also, phylogenetic relationships among CMV-satRNAs were derived from RPA data (by parsimony) or from dij (by neighbor joining). From these analyses, a model for the evolution of CMV-satRNAs in field epidemics can be built. High genetic variability of CMV-satRNA results in very heterogeneous populations, even compared with those of other RNA genomes. The high diversity of the population is maintained through time by the continuous generation of variants by mutation, counterbalanced by negative selection; this results in a certain replacement of haplotypes from year to year. The sequential accumulation of mutations in CMV-satRNA leads to fast genetic divergence to reach what appears to be an upper permitted threshold.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Fraile A., García-Arenal F. Secondary structure as a constraint on the evolution of a plant viral satellite RNA. J Mol Biol. 1991 Oct 20;221(4):1065–1069. doi: 10.1016/0022-2836(91)90916-t. [DOI] [PubMed] [Google Scholar]
- Fraile A., Moriones E., García-Arenal F. Characterization of a satellite RNA associated with strain K8 of cucumber mosaic virus. Nucleic Acids Res. 1990 Aug 11;18(15):4593–4593. doi: 10.1093/nar/18.15.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemond M., Lauquin G. J. The cDNA of cucumber mosaic virus-associated satellite RNA has in vivo biological properties. Biochem Biophys Res Commun. 1988 Feb 29;151(1):388–395. doi: 10.1016/0006-291x(88)90605-5. [DOI] [PubMed] [Google Scholar]
- Kaper J. M., Tousignant M. E. Cucumber mosaic virus-associating RNA 5. I. Role of host plant and helper strain in determining amount of associated RNA 5 with virions. Virology. 1977 Jul 1;80(1):186–195. doi: 10.1016/0042-6822(77)90391-9. [DOI] [PubMed] [Google Scholar]
- Kaper J. M., Tousignant M. E., Lot H. A low molecular weight replicating RNA associated with a divided genome plant virus: defective or satellite RNA? Biochem Biophys Res Commun. 1976 Oct 18;72(4):1237–1243. [PubMed] [Google Scholar]
- Kurath G., Palukaitis P. RNA sequence heterogeneity in natural populations of three satellite RNAs of cucumber mosaic virus. Virology. 1989 Nov;173(1):231–240. doi: 10.1016/0042-6822(89)90239-0. [DOI] [PubMed] [Google Scholar]
- Kurath G., Palukaitis P. Serial passage of infectious transcripts of a cucumber mosaic virus satellite RNA clone results in sequence heterogeneity. Virology. 1990 May;176(1):8–15. doi: 10.1016/0042-6822(90)90224-f. [DOI] [PubMed] [Google Scholar]
- Moriones E., Diaz I., Rodriguez-Cerezo E., Fraile A., Garcia-Arenal F. Differential interactions among strains of tomato aspermy virus and satellite RNAs of cucumber mosaic virus. Virology. 1992 Feb;186(2):475–480. doi: 10.1016/0042-6822(92)90012-e. [DOI] [PubMed] [Google Scholar]
- Moriones E., Fraile A., García-Arenal F. Host-associated selection of sequence variants from a satellite RNA of cucumber mosaic virus. Virology. 1991 Sep;184(1):465–468. doi: 10.1016/0042-6822(91)90871-8. [DOI] [PubMed] [Google Scholar]
- Palukaitis P., Roossinck M. J., Dietzgen R. G., Francki R. I. Cucumber mosaic virus. Adv Virus Res. 1992;41:281–348. doi: 10.1016/s0065-3527(08)60039-1. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Cerezo E., Moya A., García-Arenal F. Variability and evolution of the plant RNA virus pepper mild mottle virus. J Virol. 1989 May;63(5):2198–2203. doi: 10.1128/jvi.63.5.2198-2203.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roossinck M. J., Sleat D., Palukaitis P. Satellite RNAs of plant viruses: structures and biological effects. Microbiol Rev. 1992 Jun;56(2):265–279. doi: 10.1128/mr.56.2.265-279.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Tien P., Wu G. S. Satellite RNA for the biocontrol of plant disease. Adv Virus Res. 1991;39:321–339. doi: 10.1016/s0065-3527(08)60799-x. [DOI] [PubMed] [Google Scholar]
- Winter E., Yamamoto F., Almoguera C., Perucho M. A method to detect and characterize point mutations in transcribed genes: amplification and overexpression of the mutant c-Ki-ras allele in human tumor cells. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7575–7579. doi: 10.1073/pnas.82.22.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]