Abstract
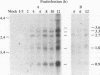
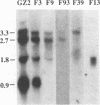
We have determined the nucleotide sequence and transcription pattern of a group of open reading frames of pseudorabies virus (PRV), which are located at the right end of the BamHI-G fragment from 0.664 to 0.695 map units in the unique long region of the genome. Nucleotide sequence analysis revealed four open reading frames. The first three correspond in genome location to the herpes simplex virus type 1 (HSV-1) open reading frames UL1, which codes for glycoprotein L (gL); UL2, which codes for a uracil-DNA glycosylase; and UL3, which codes for a polypeptide of unknown function. The fourth open reading frame, UL3.5, is not present in the HSV-1 genome. Northern (RNA) blot analysis with oligonucleotide and cDNA probes revealed four abundant mRNA species of 3.3, 2.7, 1.8, and 0.9 kb, which are likely to yield polypeptides encoded by the UL1, -2, -3, and -3.5 open reading frames, respectively. All four transcripts were of the early-late kinetic class, transcribed in the same direction, and 3' coterminal. The UL2 and UL3 genes of PRV and HSV-1 have significant amino acid sequence homology, while the UL1 genes are positional homologs and the UL3.5 gene is unique to PRV.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984 Jul 19;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T., Veach R. A., Ihara S. Localization of the regions of homology between the genomes of herpes simplex virus, type 1, and pseudorabies virus. Virology. 1983 May;127(1):194–204. doi: 10.1016/0042-6822(83)90383-5. [DOI] [PubMed] [Google Scholar]
- Chee M. S., Bankier A. T., Beck S., Bohni R., Brown C. M., Cerny R., Horsnell T., Hutchison C. A., 3rd, Kouzarides T., Martignetti J. A. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- Cheung A. K. Cloning of the latency gene and the early protein 0 gene of pseudorabies virus. J Virol. 1991 Oct;65(10):5260–5271. doi: 10.1128/jvi.65.10.5260-5271.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A. K. Detection of pseudorabies virus transcripts in trigeminal ganglia of latently infected swine. J Virol. 1989 Jul;63(7):2908–2913. doi: 10.1128/jvi.63.7.2908-2913.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A. J., Scott J. E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986 Sep;67(Pt 9):1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- Davison A. J., Wilkie N. M. Location and orientation of homologous sequences in the genomes of five herpesviruses. J Gen Virol. 1983 Sep;64(Pt 9):1927–1942. doi: 10.1099/0022-1317-64-9-1927. [DOI] [PubMed] [Google Scholar]
- DeMarchi J. M., Lu Z. Q., Rall G., Kupershmidt S., Ben-Porat T. Structural organization of the termini of the L and S components of the genome of pseudorabies virus. J Virol. 1990 Oct;64(10):4968–4977. doi: 10.1128/jvi.64.10.4968-4977.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson L., Browne H., Wargent V., Davis-Poynter N., Primorac S., Goldsmith K., Minson A. C., Johnson D. C. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J Virol. 1992 Apr;66(4):2240–2250. doi: 10.1128/jvi.66.4.2240-2250.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye J. F., Gompels U. A., Minson A. C. Glycoprotein H of human cytomegalovirus (HCMV) forms a stable complex with the HCMV UL115 gene product. J Gen Virol. 1992 Oct;73(Pt 10):2693–2698. doi: 10.1099/0022-1317-73-10-2693. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J., Dalrymple M. A., Davison A. J., Dolan A., Frame M. C., McNab D., Perry L. J., Scott J. E., Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988 Jul;69(Pt 7):1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- Moriuchi H., Moriuchi M., Smith H. A., Straus S. E., Cohen J. I. Varicella-zoster virus open reading frame 61 protein is functionally homologous to herpes simplex virus type 1 ICP0. J Virol. 1992 Dec;66(12):7303–7308. doi: 10.1128/jvi.66.12.7303-7308.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullaney J., Moss H. W., McGeoch D. J. Gene UL2 of herpes simplex virus type 1 encodes a uracil-DNA glycosylase. J Gen Virol. 1989 Feb;70(Pt 2):449–454. doi: 10.1099/0022-1317-70-2-449. [DOI] [PubMed] [Google Scholar]
- Muller S. J., Caradonna S. Isolation and characterization of a human cDNA encoding uracil-DNA glycosylase. Biochim Biophys Acta. 1991 Feb 16;1088(2):197–207. doi: 10.1016/0167-4781(91)90055-q. [DOI] [PubMed] [Google Scholar]
- Olsen L. C., Aasland R., Wittwer C. U., Krokan H. E., Helland D. E. Molecular cloning of human uracil-DNA glycosylase, a highly conserved DNA repair enzyme. EMBO J. 1989 Oct;8(10):3121–3125. doi: 10.1002/j.1460-2075.1989.tb08464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry L. J., Rixon F. J., Everett R. D., Frame M. C., McGeoch D. J. Characterization of the IE110 gene of herpes simplex virus type 1. J Gen Virol. 1986 Nov;67(Pt 11):2365–2380. doi: 10.1099/0022-1317-67-11-2365. [DOI] [PubMed] [Google Scholar]
- Roop C., Hutchinson L., Johnson D. C. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J Virol. 1993 Apr;67(4):2285–2297. doi: 10.1128/jvi.67.4.2285-2297.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Sancar G. B. DNA repair enzymes. Annu Rev Biochem. 1988;57:29–67. doi: 10.1146/annurev.bi.57.070188.000333. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney U., Hutcheon T., van de Sande J. H. Sequence analysis, expression, and conservation of Escherichia coli uracil DNA glycosylase and its gene (ung). J Biol Chem. 1988 Jun 5;263(16):7776–7784. [PubMed] [Google Scholar]
- Worrad D. M., Caradonna S. Identification of the coding sequence for herpes simplex virus uracil-DNA glycosylase. J Virol. 1988 Dec;62(12):4774–4777. doi: 10.1128/jvi.62.12.4774-4777.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wind N., Domen J., Berns A. Herpesviruses encode an unusual protein-serine/threonine kinase which is nonessential for growth in cultured cells. J Virol. 1992 Sep;66(9):5200–5209. doi: 10.1128/jvi.66.9.5200-5209.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zijl M., van der Gulden H., de Wind N., Gielkens A., Berns A. Identification of two genes in the unique short region of pseudorabies virus; comparison with herpes simplex virus and varicella-zoster virus. J Gen Virol. 1990 Aug;71(Pt 8):1747–1755. doi: 10.1099/0022-1317-71-8-1747. [DOI] [PubMed] [Google Scholar]