Abstract
A rapidly growing area of genome research is the generation of expressed sequence tags (ESTs) in which large numbers of randomly selected cDNA clones are partially sequenced. The collection of ESTs reflects the level and complexity of gene expression in the sampled tissue. To date, the majority of plant ESTs are from nonwoody plants such as Arabidopsis, Brassica, maize, and rice. Here, we present a large-scale production of ESTs from the wood-forming tissues of two poplars, Populus tremula L. × tremuloides Michx. and Populus trichocarpa ‘Trichobel.’ The 5,692 ESTs analyzed represented a total of 3,719 unique transcripts for the two cDNA libraries. Putative functions could be assigned to 2,245 of these transcripts that corresponded to 820 protein functions. Of specific interest to forest biotechnology are the 4% of ESTs involved in various processes of cell wall formation, such as lignin and cellulose synthesis, 5% similar to developmental regulators and members of known signal transduction pathways, and 2% involved in hormone biosynthesis. An additional 12% of the ESTs showed no significant similarity to any other DNA or protein sequences in existing databases. The absence of these sequences from public databases may indicate a specific role for these proteins in wood formation. The cDNA libraries and the accompanying database are valuable resources for forest research directed toward understanding the genetic control of wood formation and future endeavors to modify wood and fiber properties for industrial use.
Keywords: cambium/forestry/functional genomics/xylem/xylogenesis
In forest trees, stem diameter growth results from the activity of the vascular cambium (1, 2). Cells that originate from this meristem differentiate into water-conducting and supportive xylem (wood) and into phloem tissues that translocate photosynthate. Development of xylem and phloem involves several fundamental processes of plant growth and development, including cell division, cell expansion, formation of secondary cell walls (involving cellulose, hemicellulose, and lignin synthesis), and programmed cell death. During these developmental steps, most of the structural and chemical properties of wood and fibers are determined.
Despite the high economic value of wood, little is known about the genetic control of wood formation. Some progress in our understanding of lignin biosynthesis has been achieved in trees (3), but much of our insight comes from studies in nonwoody model systems, such as Zinnia elegans and Arabidopsis thaliana. Induction of tracheary elements in Zinnia cell cultures has resulted in the cloning of genes involved in tracheary element differentiation (4), and screening for Arabidopsis mutants aberrant in vascular development, lignin biosynthesis, and cellulose biosynthesis has yielded interesting genes for wood formation (5–9).
In annual model plants, such as Arabidopsis, however, it is a difficult task to explore the genes that are preferentially or specifically expressed during xylogenesis. Although Arabidopsis, as most annual plants, develops a true vascular cambium and small amounts of secondary xylem (ref. 10; S.R. and B.S., unpublished data; N. Chaffey, personal communication), the xylem-forming tissues cannot be easily separated from other tissues. In contrast, during active growth of trees, the vascular cambium and its developing derivatives can readily be accessed by peeling the bark. Large amounts of wood-forming tissues can then be harvested, providing a highly enriched material for investigations on the molecular biology and biochemistry of wood formation.
A relatively rapid way to obtain information about gene expression and coding sequences of uncharacterized genomes is partial sequencing of cDNAs (11). A. thaliana and rice (Oryza sativa) have become the two most important plants for genomic analyses. By systematic sequencing of expressed sequence tags (ESTs) of Arabidopsis, ≈36,000 fragments have been submitted to the EST database (dbEST) (ref. 12; http://www.ncbi.nlm.nih.gov) representing probably ≈57% of the expected 21,000 genes in this plant. Given the rapid progress in sequencing methods, the whole Arabidopsis genome can be expected to be sequenced by the year 2000. Rice has also been a subject for EST sequencing efforts (13) and by August 1998, ≈28,000 fragments were reported to dbEST. Some other plants represented in dbEST are Zea mays (2,749 sequences), Pinus taeda (1,953 sequences), Brassica napus (1,434 sequences), and Brassica campestris (966 sequences).
Here, we report on a large-scale gene discovery program in two poplar species that are important for forest biotechnology. Populus tremula L. × tremuloides Michx. was used because it is a model clone for genetic engineering, whereas Populus trichocarpa ‘Trichobel’ belongs to one of the three most important Populus species for commercial breeding. Poplar is a fast-growing tree with a relatively small genome (550 Mb) (14) that can be propagated vegetatively; furthermore, genetic maps are available (15) and extensive quantitative trait loci mapping programs are being conducted (16). In addition, a bacterial artificial chromosome library has been constructed that will enable physical mapping of ESTs. Because various genotypes of the Populus genus are readily transformed (17, 18), the function determination of EST clones can be attained by sense and antisense approaches.
There were two goals in the formation of ESTs from poplar. (i) A large-scale effort to identify genes that are related to wood formation. For this purpose, we produced 4,809 ESTs from the cambial meristem and its developing derivatives (the cambial region) of P. tremula × tremuloides. This library was expected to provide structural genes for cell wall biosynthesis, but also to be enriched in genes involved in the developmental control of xylem and phloem formation. (ii) The next goal was to ascertain whether a smaller library from a more specific tissue would generate a higher percentage of genes related to the developmental pathways of that tissue. To this end, we have produced 883 ESTs from the developing-xylem region of P. trichocarpa to enrich for genes specifically related to xylem formation.
MATERIALS AND METHODS
cDNA Libraries.
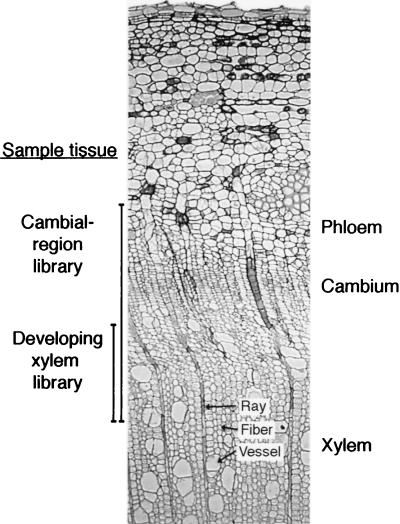
The libraries were constructed from stem tissue isolated from actively growing trees. A cambial-region library was prepared from tissues, including the developing xylem, the meristematic cambial zone, and the developing and mature phloem of P. tremula L. × tremuloides Michx. These tissues were obtained by peeling the bark and scraping both exposed surfaces with a scalpel (Fig. 1). The cDNA library was prepared from λgt22a by using the Superscript Lambda System for cDNA Synthesis and Cloning (GIBCO/BRL) and packaged into λ particles with the Gigapack II Gold (Stratagene), according to the manufacturers’ instructions. λ DNA was isolated from an aliquot of the cDNA library representing 200,000 clones, and the cDNA inserts were isolated and ligated into pBluescript SK (Stratagene). Bacterial clones of the cambial-region cDNA library were randomly picked, suspended in 100 μl of Tris/EDTA buffer, lysed, and stored at −20°C until analysis. A developing-xylem library was prepared from Populus trichocarpa ‘Trichobel.’ The tissues were obtained by peeling the bark and scraping the exposed xylem side (Fig. 1). cDNA prepared from the mRNA of these tissues was directionally cloned into a λZAPII vector (Stratagene) according to the manufacturer’s instructions. Plasmid clones of individual phages were obtained by in vivo excision.
Figure 1.
Transverse section of a poplar stem that shows the tissues used for the EST libraries. The cambial-region EST library was prepared from developing and mature phloem, the cambial meristem, and developing xylem. A separate library was prepared from the developing xylem only.
DNA Sequencing.
Sequencing of the cambial-region cDNA inserts was performed from the 5′ end by using PCR products as templates. Microtiter plates (with bacterial lysates) were loaded onto a robotic worktable, where PCRs, quality control, and sequencing reactions were performed automatically (A.H., unpublished data). PCRs were performed by using general vector primers and standard PCR protocols (19) and control of size and quality of the PCR products was performed by gel electrophoresis. The majority of the samples were analyzed by using the BigDye Terminator Cycle Sequencing kit (Perkin–Elmer-Applied Biosystems) and a biotinylated sequencing primer [universal sequencing primer (USP); Interactiva, Ulm, Germany] to allow capture and purification of generated sequencing products onto paramagnetic beads (M280-Streptavidin; Dynal, Oslo) before the samples were loaded on an ABI 377 DNA sequencer (Perkin–Elmer-Applied Biosystems). Sequencing of developing-xylem cDNAs was performed by using similar chemistry, but with automatically purified plasmids as templates in the cycle sequencing reactions.
Sequence Analysis.
Sequences were edited manually by using the pregap program in the staden package (20) to remove vector sequences and to make quality clipping in the 3′ end. The script exp2ace (S. Bergh, personal communication) was used to transform the pregap output to a Caenorhabditis elegans database (AceDB) format [Durbin, R., and Mieg, J. T. (1991) A C. elegans database. Documentation, code, and data available from anonymous FTP servers at lirmm.lirmm.fr, cele.mrc-lmb.cam.ac.uk, and ncbi.nlm.nih.gov]; AceDB was then used for storage of all information related to the sequences. Contaminants of vector sequences and rRNA in the data sets were identified and removed and sequences shorter than 100 bases were discarded. The individual ESTs were translated and searched against the GenBank nonredundant protein database by using blastx (21). Searches with nucleotide sequences by using blastn were also performed against GenBank nonredundant and EST databases. The search results were formatted by mspcrunch (22) and imported to the AceDB database. Scores >100 were considered as significant and the top-scoring genes were used to group the transcripts by their putative function (12, 23, 24).
Assembly of the individual ESTs into groups of sequences (clusters) representing unique transcripts was performed by using the TIGR assembler (25), with parameters optimized for ESTs rather than for random genomic clones. To use the alignment editor function in gap4 (26), output information regarding assembled clusters and failed alignments from the assembler were converted into gap format in two steps. (i) A Java application called tigrasm2staden (M.L., unpublished data) was used to group the assembled sequences in a flat file. The sequences that failed in the assembly process as a result of too many sequencing errors or potential alternative splicing were also added as separate readings in the corresponding flat file. (ii) The flat files were then converted to gap format by using the program convert (staden package). The gap4 program was subsequently used to manually assemble or reject the potentially overlapping sequences.
RESULTS
cDNA Sequencing. The generated cambial-region cDNA library from P. tremula × tremuloides, and the developing-xylem cDNA library from P. trichocarpa, had average insert sizes of 1.0–1.5 kb. By using semiautomatic procedures for DNA sequencing, a total of 4,883 + 928 sequences were obtained from the 5′ end of the respective libraries that reflected the genes expressed in these tissues. The average read-length after vector and quality clipping was 391 bp and 446 bp for the cambial-region and developing-xylem cDNA libraries, respectively. After vector sequences, ESTs identified as rRNA, and sequences shorter than 100 bases were removed, the final number of ESTs was 4,809 and 883 for the two libraries.
Assembly Results.
A total of 751 clusters were formed after assembly of the 4,809 ESTs from the cambial region. Each cluster consisted of at least two ESTs and was considered to be derived from the same gene by taking sequence ambiguities into account. The clusters contained a total of 2,572 sequences, whereas 2,237 sequences remained as singleton ESTs, not identical to any other EST in the data set. This corresponds to a redundancy of 53%—i.e., there is a 53% chance that a new sequence will already be represented in the data set. The number of sequences in a given cluster ranged between two (417 clusters) and 30 (1 cluster). The 883 developing-xylem ESTs were assembled into 78 clusters that contained a total of 230 sequences (26% redundancy) and 653 sequences as singleton ESTs. In this library, the number of sequences in a given cluster ranged between 2 (55 clusters) and 17 (1 cluster). Assuming that the singleton ESTs were unique transcripts, a total of 2,988 and 731 unique transcripts were found in the two respective libraries.
Similarities.
Most of the ESTs in both cDNA libraries could be assigned a cellular role on the basis of sequence similarity to proteins with known function by using a blastx stringency score >100. For the cambial-region library, 63% of the ESTs could be putatively identified compared with 54% for the developing-xylem library. The total number of proteins with known function that were identified in the two libraries was 820, taking into account that many of the unique transcripts represent isoforms of the same protein. Of these proteins, 164 were found in both libraries, 581 in the cambial-region library only, and 75 in the developing-xylem library only. A complete list of genes identified in each library is available through the Internet (http://www.biochem.kth.se/PopulusDB). The remaining ESTs had either no significant similarity to any protein or DNA sequence in the databases (12% and 9% for the cambial-region and developing-xylem libraries, respectively) or showed significant similarity to ESTs of unknown function or genomic clones from other organisms (25% and 37% for the cambial-region and developing-xylem libraries, respectively). After removal of the identified genes, 1,474 unique transcripts of unknown function remained in the two libraries, including both clusters as well as singletons. This number could be overestimated because some of the sequences could be nonoverlapping sequences of the same transcript.
The most highly abundant transcripts of the two libraries are presented in Table 1. The two libraries contained different highly abundant transcripts, and the frequencies were higher in the developing-xylem library. Several of these ESTs were highly similar (scores >500) to proteins of known function, including laccase, S-adenosyl-l-methionine synthase, elongation factor 1-α, and 14-3-3-like protein. Several other highly abundant transcripts were moderately similar (scores between 100 and 250) to cyclophilin, translationally controlled tumor protein, blue copper protein, and ADP-ribosylation factor. The EST annotated as high-mobility group (HMG) protein exhibited a low similarity to other HMG proteins (score of 94), but contained the highly conserved HMG domain (27), indicating that it probably encodes an HMG protein. The identity of the most highly abundant transcript in the developing-xylem library is ambiguous because it was similar both to nodulin, which is expressed during nodule formation in alfalfa (score 136), and to an extracellular glycoprotein in carrot (score 124). Three highly abundant transcripts in Table 1 lacked significant similarity to any protein in the databases and are denoted unknown.
Table 1.
Assembled clusters that correspond to the highest expressed genes from the cambial-region and the developing- xylem libraries
| Putative gene identification | Hits* | ‰† |
|---|---|---|
| Cambial-region ESTs | ||
| Cyclophilin | 30 | 6.2 |
| Unknown (I) | 28 | 5.8 |
| Translationally controlled tumor protein | 26 | 5.4 |
| Unknown (II) | 23 | 4.8 |
| Blue copper protein | 21 | 4.4 |
| ADP-ribosylation factor (I) | 21 | 4.4 |
| HMG protein 1 | 17 | 3.5 |
| ADP-ribosylation factor (II) | 16 | 3.3 |
| Developing-xylem ESTs | ||
| Nodulin | 17 | 19.3 |
| Laccase | 14 | 15.9 |
| Unknown | 10 | 11.3 |
| S-Adenosyl-l-methionine synthase | 8 | 9.1 |
| Elongation factor 1-α | 7 | 7.9 |
| 14-3-3-like protein | 7 | 7.9 |
Total number of ESTs in each cluster.
Frequency of EST in relation to the size of each library.
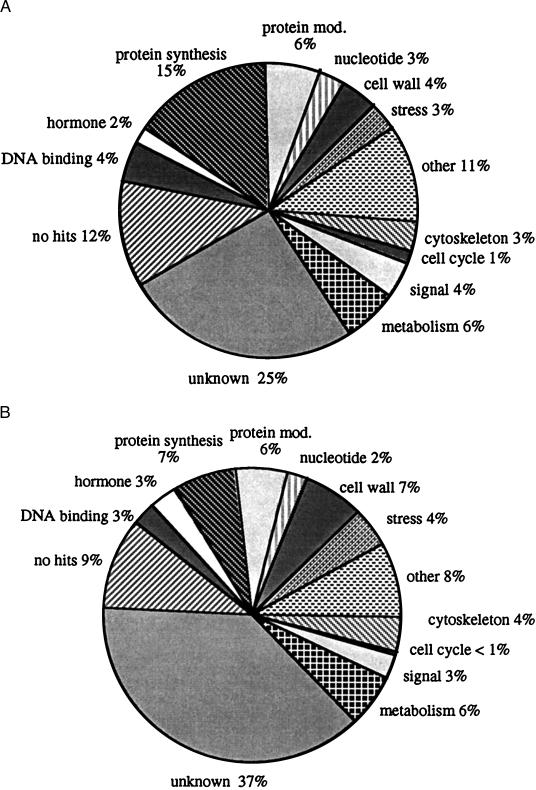
ESTs with putative protein identities were classified into 12 functional groups (Fig. 2), principally based on the catalogues established for Escherichia coli, Saccharomyces cerevisiae, and A. thaliana (12, 23, 24). In the cambial-region EST library (Fig. 2A), most of the known genes belong to functional groups related to the general housekeeping responsibilities of the cells, such as protein synthesis, general metabolism, and nucleic acid and amino acid synthesis. The genes of more direct interest to forest biotechnology were classified within the smaller functional groups. Genes involved in cell wall formation made up 4% of the total and comprised genes involved in lignin, cellulose, and hemicellulose biosynthesis. Hormone biosynthesis and related genes represented 2% of the clones, with those involved in ethylene and auxin-related pathways being the most abundant. DNA-binding proteins constituted 4% of the total and included not only histones but also several putative transcriptional regulators with similarities to both MADS box and homeobox proteins.
Figure 2.
Classification of the 4,809 and 883 ESTs from the cambial-region (A) and developing-xylem (B) libraries, respectively. ESTs with blastx scores >100 were classified into functional categories: unknown, with similarity to uncharacterized DNA or protein sequences in existing databases; no hits, no sequence similarity found in existing databases; DNA-binding proteins; hormone synthesis-related proteins; protein synthesis; protein modification, degradation, and targeting; nucleotide and amino acid metabolism; cell wall formation; stress-related proteins; other proteins whose function does not fit into the other categories; cytoskeleton; cell cycle control; signal transduction; and general metabolism.
Comparison of the two libraries revealed distinct differences in the relative distribution of the functional groups (Fig. 2B). The most significant difference was found in the expression of cell wall-related genes in the developing-xylem library, which was almost twice as high as in the cambial-region library. In addition, the expression of protein synthesis-related genes was only half as abundant in the developing-xylem library. Furthermore, whereas the developing-xylem library had more proteins of unknown function (54% vs. 37%), the cambial-region library had more cDNAs that had no similarity to sequences in existing databases (12% vs. 9%).
Genes putatively related to cell wall formation in the cambial-region and developing-xylem libraries are shown in Table 2. Lignin biosynthetic genes were present in both libraries, but were generally more abundant in the developing-xylem library. There were also distinct differences in the expression of other genes related to lignification, specifically the abundance of laccase and peroxidase. Peroxidase was more abundantly expressed in the cambial-region library, whereas laccase was highly expressed in the developing-xylem library.
Table 2.
Examples of ESTs in the two libraries with similarities to genes related to cell wall formation
| Putative gene identification | Cambial region* | Developing xylem* |
|---|---|---|
| Arabinogalactan-like protein | 7 (1.5) | 2 (2.3) |
| Caffeoyl-CoA 3-O-methyltransferase | 16 (3.3) | 7 (7.9) |
| Caffeic acid O-methyltransferase | 5 (1.0) | 2 (2.3) |
| Basic cellulase | 1 (0.2) | |
| Cellulose synthase | 10 (2.1) | 1 (1.1) |
| trans-Cinnamate 4-monooxygenase | 6 (1.2) | 1 (1.1) |
| Cinnamyl-alcohol dehydrogenase | 16 (3.3) | 3 (3.4) |
| Cinnamoyl-CoA reductase | 4 (0.8) | 3 (3.4) |
| 4-Coumarate-CoA ligase | 3 (0.6) | |
| Expansin | 10 (2.1) | 8 (9.1) |
| Extensin | 6 (1.2) | 2 (2.3) |
| Ferulate 5-hydroxylase | 2 (0.4) | 1 (1.1) |
| β-1,3-Glucanase homolog | 1 (0.2) | 1 (1.1) |
| Endo-1,4-β-glucanase | 3 (0.6) | |
| Endo-1,3-β-d-glucosidase | 1 (0.2) | |
| Exo-poly-α-d-galacturonosidase | 3 (0.6) | |
| β-d-Glucan exohydrolase | 1 (0.2) | |
| β-Glucosidase | 7 (1.5) | |
| Hydroxyproline-rich glycoprotein | 4 (0.8) | |
| Laccase | 2 (0.4) | 16 (18.1) |
| Pectate lyase | 13 (2.7) | 1 (1.1) |
| Pectin acetylesterase | 1 (0.2) | |
| Pectin methylesterase | 7 (1.5) | 2 (2.3) |
| Peroxidase, anionic | 12 (2.5) | 1 (1.1) |
| Phenylalanine ammonia lyase | 1 (0.2) | 2 (2.3) |
| Polygalacturonase | 2 (0.4) | |
| Xyloglucan endotransglycosylase | 7 (1.5) | 2 (2.3) |
| Xylan endohydrolase isoenzyme | 1 (0.2) | 1 (1.1) |
The number of ESTs for each gene function is based on individual search results, and the frequencies in relation to the size of respective libraries are given in parentheses as ‰.
DISCUSSION
The poplar EST data presented here will be valuable in identifying genes involved in the formation of secondary xylem and phloem in plants. Recently, a similar, albeit smaller, EST database was prepared from the developing-xylem of Pinus taeda (28). The established databases will certainly identify the genes controlling the building blocks of the wood fiber—i.e., those encoding proteins important for biosynthesis of lignin, cellulose, hemicellulose, and cell wall. In addition, it will reveal genes involved in the genetic control of wood development, including important traits, such as the morphology (cell length, width, and wall thickness) and chemical structure of the wood fiber, the composition of wood cell types, and the allocation of carbon to stem growth. Ultimately, this will result in biotechnological approaches to increase the value of wood and fibers grown for industrial use.
A total of 2,988 unique transcripts (2,237 appearing once and 751 twice or more) were identified in the cambial region, and the data from the developing-xylem region revealed 731 unique transcripts (653 + 78). These numbers correspond to redundancy levels of 53% and 26% for the respective libraries, indicating that many genes remain to be identified in these libraries.
Comparison of our EST sequences to public databases indicated that 63% of the cambial-region library and 54% of the developing-xylem library shared significant similarity with proteins of known function from other organisms. This percentage is similar to that of the developing-xylem EST library from Pinus taeda, which contains 1,097 EST clones of which 59% are similar to sequences of known function (28). Putative poplar homologs for 820 proteins with known function from other organisms were identified. Within the ESTs that could be assigned a putative function, several were of specific interest to forest biotechnology. For example, ESTs corresponding to all the identified enzymes involved in the formation of the lignin monomers were identified, as were others involved in cellulose synthesis, including putative homologs of cellulose synthase (Table 2). In addition, both libraries contained many ESTs that may encode proteins involved in cell wall expansion as well as cross-linking and modifying the fiber structure and composition. Potential developmental regulators of wood formation were also identified, such as proteins related to hormone synthesis and perception, cell cycle control, and putative transcriptional activators with similarity to MADS box and homeodomain proteins.
Of particular interest is the set of 12% of the ESTs in the cambial-region library, and 9% of the ESTs in the developing-xylem library, that showed no sequence similarity to any sequences in the databases (Fig. 2). By considering the large number of ESTs and genomic sequences available for Arabidopsis and rice, many of these no-hit ESTs could probably represent genes specifically involved in the formation of secondary tissues. Future functional analysis of these gene products may reveal novel control mechanisms and previously unknown details of the biosynthetic pathways for wood formation.
The smaller developing-xylem library was sequenced to determine whether a more specific EST library would identify a higher proportion of genes specifically related to xylogenesis. Whereas the number of genes involved in most housekeeping functions was essentially the same in the two libraries, there was a marked increase in the proportion of genes related to cell wall formation in the developing-xylem library (Fig. 2) as well as a concomitant decrease in the proportion of protein synthesis-related genes. This observation indicates a shift in the metabolism of developing-xylem cells away from primary metabolism toward cell wall formation. In a specific comparison between genes related to cell wall formation in the two libraries, it was evident that most of the transcripts coding for enzymes involved in the synthesis of lignin were present at a higher frequency in the developing-xylem library (Table 2). Therefore, more specific libraries are interesting not only because they are enriched in the genes responsible for the various processes of the tissue but also because they may reveal subtle differences in gene expression patterns of related genes, as indicated by the differential expression of lignin biosynthesis genes in this study. To extend the utility of specific libraries, we will focus on the production of ESTs from distinct zones of developing xylem to better understand the stages of wood formation, including cell division, cell expansion, secondary wall formation, and programmed cell death.
The EST approach to gene discovery described here gives an indication of the level of expression of each gene. Therefore, it is interesting to investigate more thoroughly the most highly expressed genes to gain an understanding of the most active pathways in the sampled tissues (Table 1). Several of the highly abundant ESTs are similar to genes that encode housekeeping proteins. These ESTs include cyclophilin, ADP-ribosylation factor, HMG-1, 14-3-3-like protein, and elongation factor 1-α. Two genes expressed in the developing-xylem library have been implicated in lignin biosynthesis, laccase and S-adenosyl-l-methionine synthase. Laccase is a blue copper oxidase involved in monolignol polymerization (29, 30). The sequence in the developing-xylem library showed the highest similarity to a laccase from Liriodendron tulipifera (yellow poplar; GenBank accession U73106), but also to a laccase from Acer pseudoplatanus that has been demonstrated to polymerize monolignol precursors in vitro (31). It is remarkable that the abundance of the laccase in the developing-xylem library is 45-fold higher than that of the cambial-region library, whereas the abundance of peroxidase, another enzyme believed to be involved in polymerization of monolignols, is more abundant in the cambial-region library (Table 2). A higher proportion of laccase compared with peroxidase ESTs was also found in the developing-xylem library from Pinus taeda (28), suggesting that laccase polymerization of monolignols in the xylem might be important for both angiosperm and gymnosperm trees.
S-Adenosyl-l-methionine is a highly abundant transcript in the developing-xylem library (Table 1) and, together with six other clusters that contain two to three members each, accounts for 2.5% of the ESTs in this library (data not shown). S-Adenosyl-l-methionine synthase is a universal methyl group donor in transmethylation reactions that involve many types of acceptor molecules (32). S-Adenosyl-l-methionine synthase plays a role in the methylation of monolignol precursors during lignin biosynthesis and has been found to be specifically expressed in vascular tissues of Arabidopsis (33). In addition, it is coordinately induced with bispecific caffeic acid/5-hydroxyferulic acid-O-methyltransferase by fungal elicitors in alfalfa (34). In the developing-xylem library from Pinus taeda, methionine synthase, which is also involved in methyl transfer, was a highly abundant transcript that accounted for >1% of the pine ESTs. These observations provide evidence for a strong requirement for methyl groups in lignin formation in both angiosperms and gymnosperms (28).
Another putative lignin biosynthesis gene from the abundant ESTs in the cambial-region library is the blue copper protein (Table 1). This sequence shows the highest similarity to a pea pod cDNA that is specifically expressed in the lignified endocarp and is suggested to be involved in oxidative polymerization reactions of lignin monomers (35). Furthermore, the most highly expressed gene in the developing-xylem library may code for a cell wall protein. This cluster shows the highest similarity to an alfalfa ENOD8 gene that functions in nodule structure formation (36). This EST also shows high similarity to a glycoprotein localized to cell walls in carrot suspension cells (37).
The functions of the remaining highly abundant ESTs are uncertain (Table 1). The two “unknown” clusters from the cambial-region ESTs show weak similarity to ag13 of Alnus glutinosa, which is expressed during Frankia infection (38), but the repetitive nature of the sequences indicates a low statistical significance of these hits. The nucleotide sequences of the two clusters, however, are ≈80% identical, suggesting that they encode proteins of similar function. The unknown cluster in the developing-xylem library shows no significant similarity to any protein or DNA sequence in existing databases. Further, the sequence denoted translationally controlled tumor protein in the cambial-region library has been found in many other organisms, including plants (39), but its function is unknown. The high level of expression of these genes with unknown function emphasizes how little we understand about the molecular controls of cambial growth and wood formation. Also, as more is known about the genes expressed during wood formation, the functions of many proteins may have to be redefined to include specific roles in the wood formation process.
A preliminary comparison between the EST databases from wood-forming tissues of poplar and pine has revealed several similarities. For example, the proportion of ESTs of unknown function, the enrichment of cell wall-related ESTs in the developing-xylem libraries, and the relative abundance of specific ESTs (laccase, peroxidase, and methyl donors). Thus, the molecular control of wood formation seems to have much in common between angiosperms and gymnosperms, despite the differences in wood structure and lignin composition. Investigations on the parallels in sequence and function of proteins from hardwoods and conifers will be of great interest.
Future efforts to systematically characterize the genes involved in wood formation will, in addition to conventional cDNA sequencing, include several approaches such as microarray technologies (40) and tag-sequencing of cDNAs by pyrosequencing (41). The latter is a newly developed tag-sequencing technique that allows a much higher throughput of ESTs and facilitates expression profiling and identification of tissue-specific genes.
Acknowledgments
We thank Ross Whetten and Ron Sederoff for communicating unpublished results, Jean-Charles Leplé for preparation of the P. trichocarpa cDNA library, Staffan Bergh for computational guidance, Anna Hammarsten, Helena Rönning, Ulla Wrethagen, and Caroline Buysschaert for help with the DNA sequencing, Thomas Hiltonen for help with annotations, and Martine De Cock for help with the manuscript. This work was supported by the Foundation for Strategic Research, the Knut and Alice Wallenbergs Foundation, the Swedish Agricultural Research Council, the Kempe Foundation, and the Flemish Government (IBW/3/1998). A.R. is a Research Assistant of the Fund for Scientific Research-Flanders.
ABBREVIATIONS
- EST
expressed sequence tag
- HMG
high-mobility group
Footnotes
References
- 1. Larson P R. The Vascular Cambium: Development and Structure, Springer Series in Wood Science. Berlin: Springer; 1994. [Google Scholar]
- 2.Telewski F W, Aloni R, Sauter J J. In: Biology of Populus and Its Implications for Management and Conservation. Stettler R F, Bradshaw H D Jr, Heilman P E, Hinckley T M, editors. Ottawa, ON, Canada: NRC Research; 1996. pp. 301–330. [Google Scholar]
- 3.Baucher M, Monties B, Van Montagu M, Boerjan W. Crit Rev Plant Sci. 1998;17:125–197. [Google Scholar]
- 4.Fukuda H. Plant Cell. 1997;9:1147–1156. doi: 10.1105/tpc.9.7.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arioli T, Peng L, Betzner A S, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R, et al. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- 6.Meyer K, Cusumano J C, Somerville C, Chapple C C S. Proc Natl Acad Sci USA. 1996;93:6869–6874. doi: 10.1073/pnas.93.14.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pear J R, Kawagoe Y, Schreckengost W E, Delmer D P, Stalker D M. Proc Natl Acad Sci USA. 1996;93:12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner S R, Somerville C R. Plant Cell. 1997;9:689–701. doi: 10.1105/tpc.9.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong R, Taylor J J, Ye Z-H. Plant Cell. 1997;9:2159–2170. doi: 10.1105/tpc.9.12.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolan L, Roberts K. New Phytol. 1995;131:121–128. doi: 10.1111/j.1469-8137.1995.tb03061.x. [DOI] [PubMed] [Google Scholar]
- 11.Adams M D, Kelley J M, Gocayne J D, Dubnick M, Polymeropoulos M H, Xiao H, Merril C R, Wu A, Olde B, Moreno R F, et al. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- 12.Bevan M, Bancroft I, Bent E, Love K, Piffanelli P, Goodman H, Dean C, Bergkamp R, Dirkse W, Van Staveren M, et al. Nature (London) 1998;391:485–488. doi: 10.1038/35140. [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto K, Sasaki T. Plant Mol Biol. 1997;35:135–144. [PubMed] [Google Scholar]
- 14.Bradshaw H D, Jr, Stettler R F. Theor Appl Genet. 1993;86:301–307. doi: 10.1007/BF00222092. [DOI] [PubMed] [Google Scholar]
- 15.Bradshaw H D, Jr, Villar M, Watson B D, Otto K G, Stewart S, Stettler R F. Theor Appl Genet. 1994;89:167–178. doi: 10.1007/BF00225137. [DOI] [PubMed] [Google Scholar]
- 16.Bradshaw H D, Jr, Stettler R F. Genetics. 1995;139:963–973. doi: 10.1093/genetics/139.2.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim M-S, Klopfenstein N B, Chun Y W. In: Micropropagation, Genetic Engineering, and Molecular Biology of Populus, General Technical Report RM-GTR-297. Klopfenstein N B, Chun Y W, Kim M-S, Ahuja M R, editors. Fort Collins, CO: Rocky Mountain Forest and Range Exp. Sta.; 1997. pp. 51–59. [Google Scholar]
- 18.Charest P J, Devantier Y, Jones C, Sellmer J C, McCown B H, Ellis D D. In: Micropropagation, Genetic Engineering, and Molecular Biology of Populus, (General Technical Report RM-GTR-297) Klopfenstein N B, Chun Y W, Kim M-S, Ahuja M R, editors. Fort Collins, CO: Rocky Mountain Forest and Range Exp. Sta.; 1997. pp. 60–64. [Google Scholar]
- 19.Hultman T, Bergh S, Moks T, Uhlén M. BioTechniques. 1991;10:84–93. [PubMed] [Google Scholar]
- 20.Staden R. Mol Biotechnol. 1996;5:233–241. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- 21.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Sonnhammer E L L, Durbin R. Comput Appl Biosci. 1994;10:301–307. doi: 10.1093/bioinformatics/10.3.301. [DOI] [PubMed] [Google Scholar]
- 23.Riley M. Microbiol Rev. 1993;57:862–952. doi: 10.1128/mr.57.4.862-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mewes, H. W., Albermann, K., Bähr, M., Frishman, D., Gleissner, A., Hani, J., Heumann, K., Kleine, K., Maieri, A., Oliver, S. G., et al. (1997) Nature (London) 387, Suppl., 7–65. [DOI] [PubMed]
- 25.Sutton G G, White O, Adams M D, Kervalage A R. Genome Sci Technol. 1995;1:9–19. [Google Scholar]
- 26.Bonfield J K, Smith K F, Staden R. Nucleic Acids Res. 1995;23:4992–4999. doi: 10.1093/nar/23.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grasser K D, Wohlfarth T, Bäumlein H, Feix G. Plant Mol Biol. 1993;23:619–625. doi: 10.1007/BF00019309. [DOI] [PubMed] [Google Scholar]
- 28.Allona I, Quinn M, Shoop E, Swope K, St. Cyr S, Carlis J, Riedl J, Retzel E, Campbell M M, Sederoff R, Whetten R W. Proc Natl Acad Sci USA. 1998;95:9693–9698. doi: 10.1073/pnas.95.16.9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Malley D M, Whetten R, Bao W, Chen C-L, Sederoff R R. Plant J. 1993;4:751–757. [Google Scholar]
- 30.Bao W, O’Malley D M, Whetten R, Sederoff R R. Science. 1993;260:672–674. doi: 10.1126/science.260.5108.672. [DOI] [PubMed] [Google Scholar]
- 31.LaFayette P R, Eriksson K-E L, Dean J F D. Plant Physiol. 1995;107:667–668. doi: 10.1104/pp.107.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabor C W, Tabor H. Adv Enzymol. 1984;56:251–282. doi: 10.1002/9780470123027.ch4. [DOI] [PubMed] [Google Scholar]
- 33.Peleman J, Boerjan W, Engler G, Seurinck J, Botterman J, Alliotte T, Van Montagu M, Inzé D. Plant Cell. 1989;1:81–93. doi: 10.1105/tpc.1.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gowri G, Bugos R C, Campbell W H, Maxwell C A, Dixon R A. Plant Physiol. 1991;97:7–14. doi: 10.1104/pp.97.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drew J E, Gatehouse J A. J Exp Bot. 1994;45:1873–1884. [Google Scholar]
- 36.Liu C, Yeung A T, Dickstein R. Plant Physiol. 1998;117:1127. [Google Scholar]
- 37.van Engelen F A, de Jong A J, Meijer E A, Kuil C W, Meyboom J K, Dirkse W G, Booij H, Hartog M V, Vandekerckhove J, de Vries S C, et al. Plant Mol Biol. 1995;27:901–910. doi: 10.1007/BF00037018. [DOI] [PubMed] [Google Scholar]
- 38.Guan C, Akkermans A D L, van Kammen A, Bisseling T, Pawlowski K. Physiol Plant. 1997;99:601–607. [Google Scholar]
- 39.Pay A, Heberle-Bors E, Hirt H. Plant Mol Biol. 1992;19:501–503. doi: 10.1007/BF00023399. [DOI] [PubMed] [Google Scholar]
- 40.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 41.Ronaghi M, Uhlén M, Nyrén P. Science. 1998;281:363–365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]