Abstract
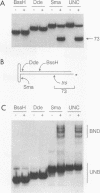
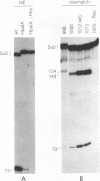
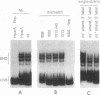
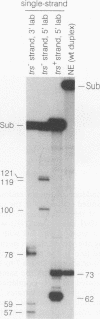
We previously demonstrated that the adeno-associated virus (AAV) Rep68 and Rep78 proteins are able to nick the AAV origin of DNA replication at the terminal resolution site (trs) in an ATP-dependent manner. Using four types of modified or mutant substrates, we now have investigated the substrate requirements of Rep68 in the trs endonuclease reaction. In the first kind of substrate, portions of the hairpinned AAV terminal repeat were deleted. Only deletions that retained virtually all of the small internal palindromes of the AAV terminal repeat were active in the endonuclease reaction. This result confirmed previous genetic and biochemical evidence that the secondary structure of the terminal repeat was an important feature for substrate recognition. In the second type of substrate, the trs was moved eight bases further away from the end of the genome. The mutant was nicked at a 50-fold-lower frequency relative to a wild-type origin, and the nick occurred at the correct trs sequence despite its new position. This finding indicated that the endonuclease reaction required a specific sequence at the trs in addition to the correct secondary structure. It also suggested that the minimum trs recognition sequence extended three bases from the cut site in the 3' direction. The third type of substrate harbored mismatched base pairs at the trs. The mismatch substrates contained a wild-type sequence on the strand normally cut but an incorrect sequence on the complementary strand. All of the mismatch mutants were capable of being nicked in the presence of ATP. However, there was substantial variation in the level of activity, suggesting that the sequence on the opposite strand may also be recognized during nicking. Analysis of the mismatch mutants also suggested that a single-stranded trs was a viable substrate for the enzyme. This interpretation was confirmed by analysis of the fourth type of substrate tested, which contained a single-stranded trs. This substrate was also cleaved efficiently by the enzyme provided that the correct strand was present in the substrate. In addition, the single-stranded substrate no longer required ATP as a cofactor for nicking. Finally, all of the substrates with mutant trss bound the Rep protein as efficiently as the wild-type did. This finding indicated that the sequence at the cut site was not involved in recognition of the terminal repeat for specific binding by the enzyme. We concluded that substrate recognition by the AAV Rep protein involves at least two and possibly as many as four features of the AAV terminal repeat.(ABSTRACT TRUNCATED AT 400 WORDS)
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ashktorab H., Srivastava A. Identification of nuclear proteins that specifically interact with adeno-associated virus type 2 inverted terminal repeat hairpin DNA. J Virol. 1989 Jul;63(7):3034–3039. doi: 10.1128/jvi.63.7.3034-3039.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns K. I., Hauswirth W. W. Adeno-associated viruses. Adv Virus Res. 1979;25:407–449. doi: 10.1016/s0065-3527(08)60574-6. [DOI] [PubMed] [Google Scholar]
- Bohenzky R. A., Berns K. I. Interactions between the termini of adeno-associated virus DNA. J Mol Biol. 1989 Mar 5;206(1):91–100. doi: 10.1016/0022-2836(89)90526-3. [DOI] [PubMed] [Google Scholar]
- Bohenzky R. A., LeFebvre R. B., Berns K. I. Sequence and symmetry requirements within the internal palindromic sequences of the adeno-associated virus terminal repeat. Virology. 1988 Oct;166(2):316–327. doi: 10.1016/0042-6822(88)90502-8. [DOI] [PubMed] [Google Scholar]
- Hong G., Ward P., Berns K. I. In vitro replication of adeno-associated virus DNA. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4673–4677. doi: 10.1073/pnas.89.10.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im D. S., Muzyczka N. Factors that bind to adeno-associated virus terminal repeats. J Virol. 1989 Jul;63(7):3095–3104. doi: 10.1128/jvi.63.7.3095-3104.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im D. S., Muzyczka N. Partial purification of adeno-associated virus Rep78, Rep52, and Rep40 and their biochemical characterization. J Virol. 1992 Feb;66(2):1119–1128. doi: 10.1128/jvi.66.2.1119-1128.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im D. S., Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990 May 4;61(3):447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- Lefebvre R. B., Riva S., Berns K. I. Conformation takes precedence over sequence in adeno-associated virus DNA replication. Mol Cell Biol. 1984 Jul;4(7):1416–1419. doi: 10.1128/mcb.4.7.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusby E., Fife K. H., Berns K. I. Nucleotide sequence of the inverted terminal repetition in adeno-associated virus DNA. J Virol. 1980 May;34(2):402–409. doi: 10.1128/jvi.34.2.402-409.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McCarty D. M., Ni T. H., Muzyczka N. Analysis of mutations in adeno-associated virus Rep protein in vivo and in vitro. J Virol. 1992 Jul;66(7):4050–4057. doi: 10.1128/jvi.66.7.4050-4057.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens R. A., Carter B. J. In vitro resolution of adeno-associated virus DNA hairpin termini by wild-type Rep protein is inhibited by a dominant-negative mutant of rep. J Virol. 1992 Feb;66(2):1236–1240. doi: 10.1128/jvi.66.2.1236-1240.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samulski R. J., Chang L. S., Shenk T. A recombinant plasmid from which an infectious adeno-associated virus genome can be excised in vitro and its use to study viral replication. J Virol. 1987 Oct;61(10):3096–3101. doi: 10.1128/jvi.61.10.3096-3101.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samulski R. J., Srivastava A., Berns K. I., Muzyczka N. Rescue of adeno-associated virus from recombinant plasmids: gene correction within the terminal repeats of AAV. Cell. 1983 May;33(1):135–143. doi: 10.1016/0092-8674(83)90342-2. [DOI] [PubMed] [Google Scholar]
- Snyder R. O., Im D. S., Muzyczka N. Evidence for covalent attachment of the adeno-associated virus (AAV) rep protein to the ends of the AAV genome. J Virol. 1990 Dec;64(12):6204–6213. doi: 10.1128/jvi.64.12.6204-6213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder R. O., Samulski R. J., Muzyczka N. In vitro resolution of covalently joined AAV chromosome ends. Cell. 1990 Jan 12;60(1):105–113. doi: 10.1016/0092-8674(90)90720-y. [DOI] [PubMed] [Google Scholar]
- Straus S. E., Sebring E. D., Rose J. A. Concatemers of alternating plus and minus strands are intermediates in adenovirus-associated virus DNA synthesis. Proc Natl Acad Sci U S A. 1976 Mar;73(3):742–746. doi: 10.1073/pnas.73.3.742. [DOI] [PMC free article] [PubMed] [Google Scholar]