Abstract
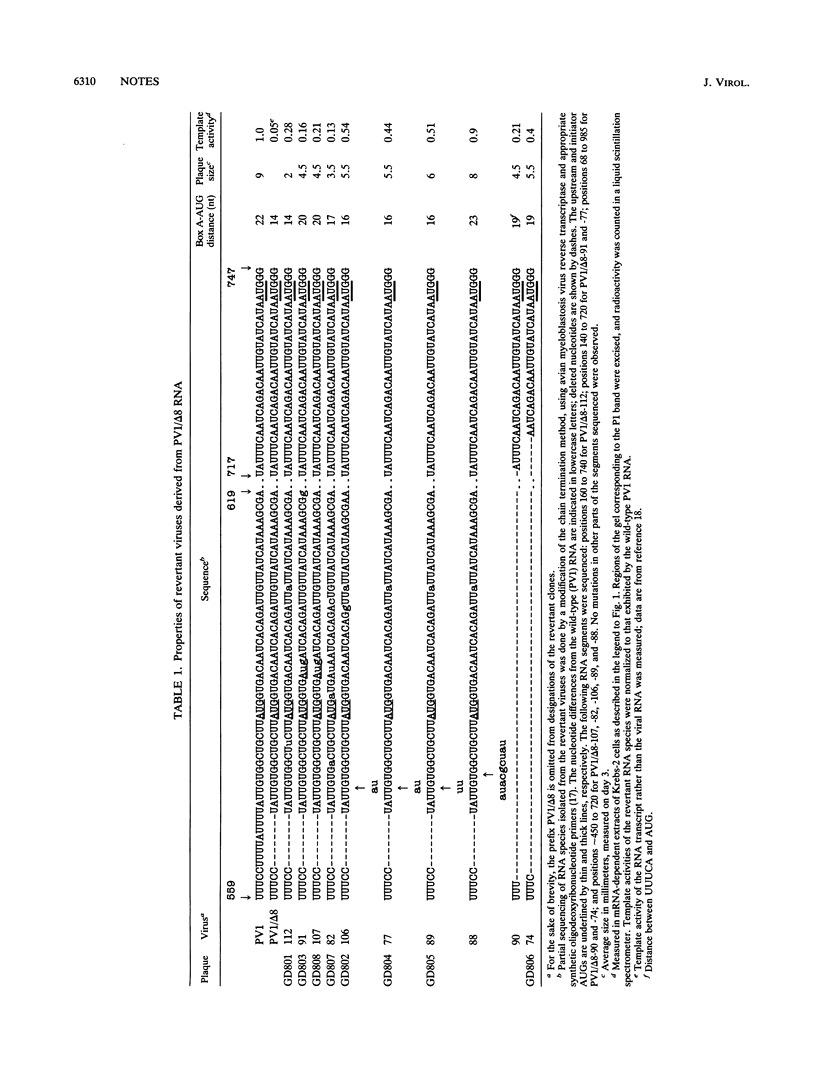
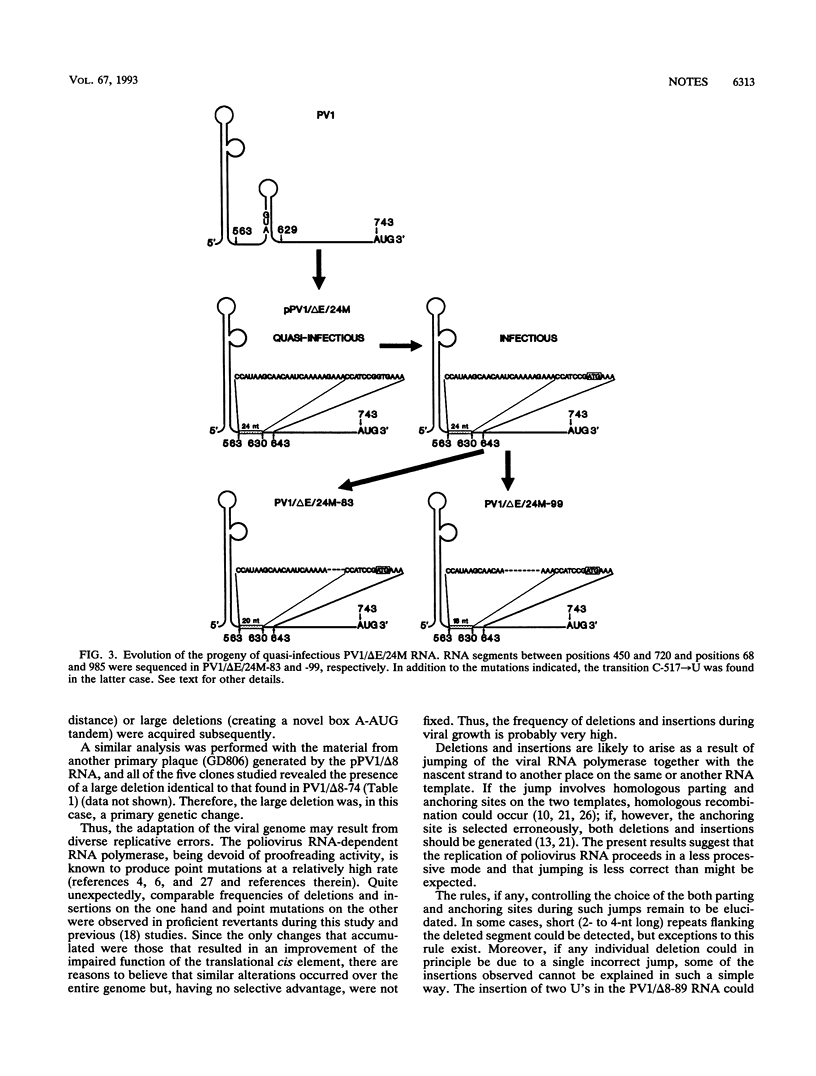
Poliovirus RNA species with nucleotides 564 to 571 deleted or with a secondary structure domain (positions 564 to 629) replaced by a shorter irregular oligonucleotide have been engineered previously; these RNAs have been considered quasi-infectious (yielding a single late revertant plaque) and dead, respectively (E. Pilipenko, A. Gmyl, Y. Svitkin, S. Maslova, A. Sinyakov, and V. Agol, Cell 68:119-131, 1992). By using large amounts of these RNAs for transfections, revertant clones with a great variety of genetic changes (point mutations, insertions of foreign sequences, short or extended deletions) were isolated. The pattern of these changes supported the notion that an appropriately spaced oligopyrimidine-AUG tandem is important for efficient poliovirus RNA translation. Structural features within and around this tandem modulated the initiation efficiency. The functional and genetic plasticities of the poliovirus genome are briefly discussed.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agol V. I. Genetic stability and instability of the cis-acting control element of the 5' untranslated region of the poliovirus RNA. Dev Biol Stand. 1993;78:11–16. [PubMed] [Google Scholar]
- Agol V. I. The 5'-untranslated region of picornaviral genomes. Adv Virus Res. 1991;40:103–180. doi: 10.1016/S0065-3527(08)60278-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman A., Jackson R. J. Initiation of translation of human rhinovirus RNA: mapping the internal ribosome entry site. Virology. 1992 Jun;188(2):685–696. doi: 10.1016/0042-6822(92)90523-r. [DOI] [PubMed] [Google Scholar]
- Haller A. A., Semler B. L. Linker scanning mutagenesis of the internal ribosome entry site of poliovirus RNA. J Virol. 1992 Aug;66(8):5075–5086. doi: 10.1128/jvi.66.8.5075-5086.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland J. J., De La Torre J. C., Steinhauer D. A. RNA virus populations as quasispecies. Curr Top Microbiol Immunol. 1992;176:1–20. doi: 10.1007/978-3-642-77011-1_1. [DOI] [PubMed] [Google Scholar]
- Iizuka N., Kohara M., Hagino-Yamagishi K., Abe S., Komatsu T., Tago K., Arita M., Nomoto A. Construction of less neurovirulent polioviruses by introducing deletions into the 5' noncoding sequence of the genome. J Virol. 1989 Dec;63(12):5354–5363. doi: 10.1128/jvi.63.12.5354-5363.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R. J., Howell M. T., Kaminski A. The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem Sci. 1990 Dec;15(12):477–483. doi: 10.1016/0968-0004(90)90302-r. [DOI] [PubMed] [Google Scholar]
- Jang S. K., Pestova T. V., Hellen C. U., Witherell G. W., Wimmer E. Cap-independent translation of picornavirus RNAs: structure and function of the internal ribosomal entry site. Enzyme. 1990;44(1-4):292–309. doi: 10.1159/000468766. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K., Baltimore D. The mechanism of RNA recombination in poliovirus. Cell. 1986 Nov 7;47(3):433–443. doi: 10.1016/0092-8674(86)90600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura N., Semler B. L., Rothberg P. G., Larsen G. R., Adler C. J., Dorner A. J., Emini E. A., Hanecak R., Lee J. J., van der Werf S. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature. 1981 Jun 18;291(5816):547–553. doi: 10.1038/291547a0. [DOI] [PubMed] [Google Scholar]
- Kuge S., Nomoto A. Construction of viable deletion and insertion mutants of the Sabin strain of type 1 poliovirus: function of the 5' noncoding sequence in viral replication. J Virol. 1987 May;61(5):1478–1487. doi: 10.1128/jvi.61.5.1478-1487.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge S., Saito I., Nomoto A. Primary structure of poliovirus defective-interfering particle genomes and possible generation mechanisms of the particles. J Mol Biol. 1986 Dec 5;192(3):473–487. doi: 10.1016/0022-2836(86)90270-6. [DOI] [PubMed] [Google Scholar]
- Nicholson R., Pelletier J., Le S. Y., Sonenberg N. Structural and functional analysis of the ribosome landing pad of poliovirus type 2: in vivo translation studies. J Virol. 1991 Nov;65(11):5886–5894. doi: 10.1128/jvi.65.11.5886-5894.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomoto A., Omata T., Toyoda H., Kuge S., Horie H., Kataoka Y., Genba Y., Nakano Y., Imura N. Complete nucleotide sequence of the attenuated poliovirus Sabin 1 strain genome. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5793–5797. doi: 10.1073/pnas.79.19.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T. V., Hellen C. U., Wimmer E. Translation of poliovirus RNA: role of an essential cis-acting oligopyrimidine element within the 5' nontranslated region and involvement of a cellular 57-kilodalton protein. J Virol. 1991 Nov;65(11):6194–6204. doi: 10.1128/jvi.65.11.6194-6204.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko E. V., Blinov V. M., Romanova L. I., Sinyakov A. N., Maslova S. V., Agol V. I. Conserved structural domains in the 5'-untranslated region of picornaviral genomes: an analysis of the segment controlling translation and neurovirulence. Virology. 1989 Feb;168(2):201–209. doi: 10.1016/0042-6822(89)90259-6. [DOI] [PubMed] [Google Scholar]
- Pilipenko E. V., Gmyl A. P., Maslova S. V., Svitkin Y. V., Sinyakov A. N., Agol V. I. Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell. 1992 Jan 10;68(1):119–131. doi: 10.1016/0092-8674(92)90211-t. [DOI] [PubMed] [Google Scholar]
- Pilipenko E. V., Maslova S. V., Sinyakov A. N., Agol V. I. Towards identification of cis-acting elements involved in the replication of enterovirus and rhinovirus RNAs: a proposal for the existence of tRNA-like terminal structures. Nucleic Acids Res. 1992 Apr 11;20(7):1739–1745. doi: 10.1093/nar/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racaniello V. R., Baltimore D. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4887–4891. doi: 10.1073/pnas.78.8.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova L. I., Blinov V. M., Tolskaya E. A., Viktorova E. G., Kolesnikova M. S., Guseva E. A., Agol V. I. The primary structure of crossover regions of intertypic poliovirus recombinants: a model of recombination between RNA genomes. Virology. 1986 Nov;155(1):202–213. doi: 10.1016/0042-6822(86)90180-7. [DOI] [PubMed] [Google Scholar]
- Stanway G., Cann A. J., Hauptmann R., Hughes P., Clarke L. D., Mountford R. C., Minor P. D., Schild G. C., Almond J. W. The nucleotide sequence of poliovirus type 3 leon 12 a1b: comparison with poliovirus type 1. Nucleic Acids Res. 1983 Aug 25;11(16):5629–5643. doi: 10.1093/nar/11.16.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin Y. V., Maslova S. V., Agol V. I. The genomes of attenuated and virulent poliovirus strains differ in their in vitro translation efficiencies. Virology. 1985 Dec;147(2):243–252. doi: 10.1016/0042-6822(85)90127-8. [DOI] [PubMed] [Google Scholar]
- Teterina N. L., Kean K. M., Gorbalenya A. E., Agol V. I., Girard M. Analysis of the functional significance of amino acid residues in the putative NTP-binding pattern of the poliovirus 2C protein. J Gen Virol. 1992 Aug;73(Pt 8):1977–1986. doi: 10.1099/0022-1317-73-8-1977. [DOI] [PubMed] [Google Scholar]
- Tolskaya E. A., Ivannikova T. A., Kolesnikova M. S., Drozdov S. G., Agol V. I. Postinfection treatment with antiviral serum results in survival of neural cells productively infected with virulent poliovirus. J Virol. 1992 Aug;66(8):5152–5156. doi: 10.1128/jvi.66.8.5152-5156.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolskaya E. A., Romanova L. I., Blinov V. M., Viktorova E. G., Sinyakov A. N., Kolesnikova M. S., Agol V. I. Studies on the recombination between RNA genomes of poliovirus: the primary structure and nonrandom distribution of crossover regions in the genomes of intertypic poliovirus recombinants. Virology. 1987 Nov;161(1):54–61. doi: 10.1016/0042-6822(87)90170-x. [DOI] [PubMed] [Google Scholar]
- Ward C. D., Flanegan J. B. Determination of the poliovirus RNA polymerase error frequency at eight sites in the viral genome. J Virol. 1992 Jun;66(6):3784–3793. doi: 10.1128/jvi.66.6.3784-3793.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre J. C., Giachetti C., Semler B. L., Holland J. J. High frequency of single-base transitions and extreme frequency of precise multiple-base reversion mutations in poliovirus. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2531–2535. doi: 10.1073/pnas.89.7.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]