Abstract
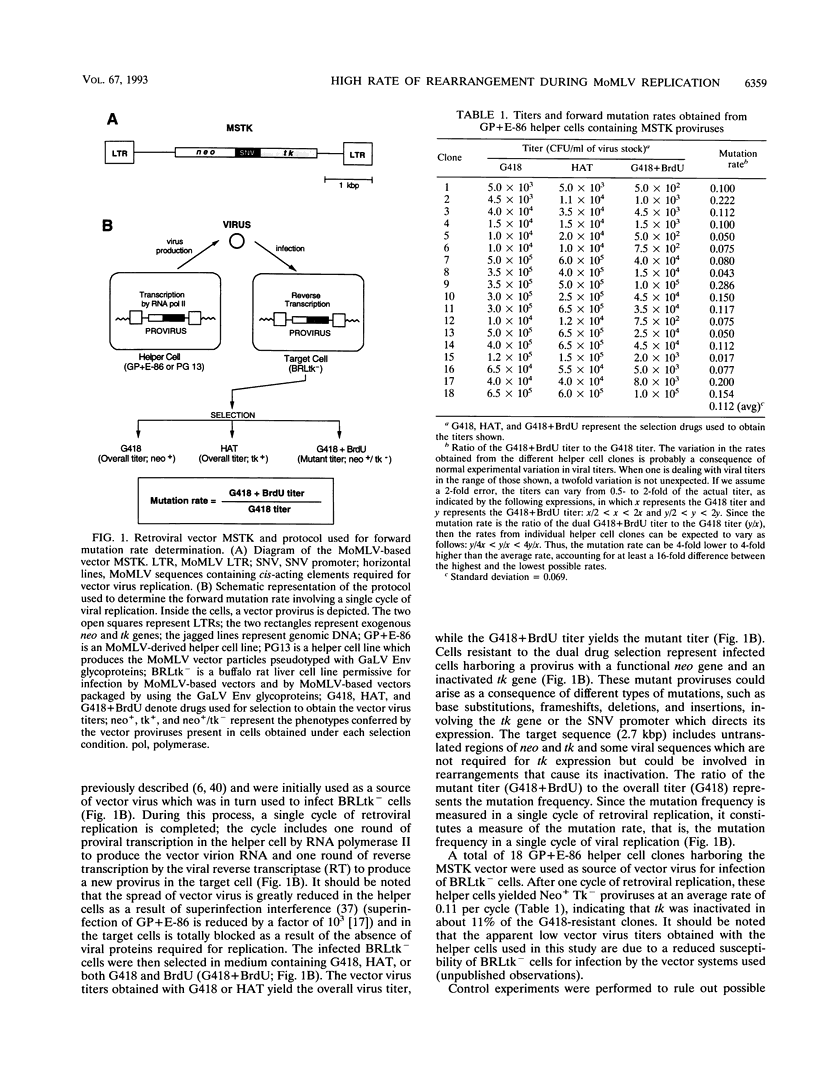
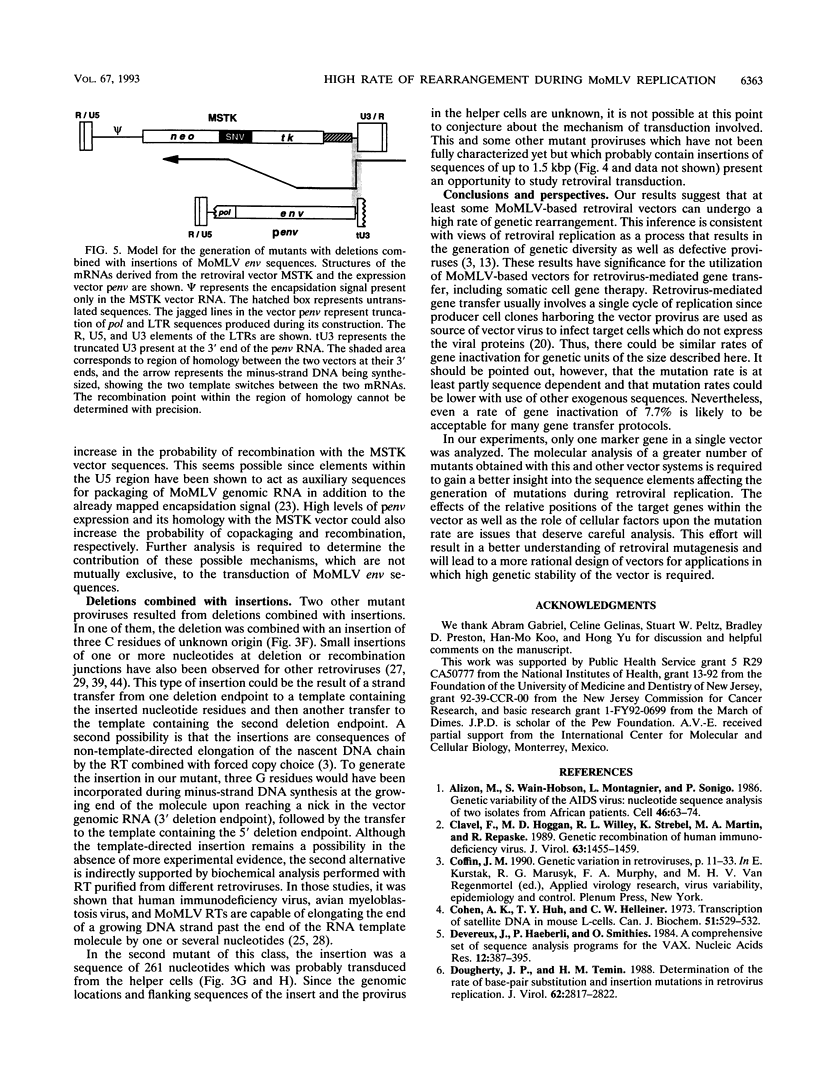
A protocol was designed to measure the forward mutation rate over an entire gene replicated as part of a Moloney murine leukemia virus-based vector. For these studies, the herpes simplex virus thymidine kinase (tk) gene under the control of the spleen necrosis virus U3 promoter was used as target sequence since it allows selection for either the functional or the inactivated gene. Our results indicate that after one round of retroviral replication, the tk gene is inactivated at an average rate of 0.08 per cycle of replication. Southern blotting revealed that the majority of the mutant proviruses resulted from gross rearrangements and that deletions of spleen necrosis virus and tk sequences were the most frequent cause of the gene inactivation. Sequence analysis of the mutant proviruses suggested that homologous as well as nonhomologous recombination was involved in the observed rearrangements. Some mutations consisted of simple deletions, and others consisted of deletions combined with insertions. The frequency at which these mutations occurred during one cycle of retroviral replication provides evidence indicating that Moloney murine leukemia virus-based vectors may undergo genetic rearrangement at high rates. The high rate of rearrangement and its relevance for retrovirus-mediated gene transfer are discussed.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alizon M., Wain-Hobson S., Montagnier L., Sonigo P. Genetic variability of the AIDS virus: nucleotide sequence analysis of two isolates from African patients. Cell. 1986 Jul 4;46(1):63–74. doi: 10.1016/0092-8674(86)90860-3. [DOI] [PubMed] [Google Scholar]
- Clavel F., Hoggan M. D., Willey R. L., Strebel K., Martin M. A., Repaske R. Genetic recombination of human immunodeficiency virus. J Virol. 1989 Mar;63(3):1455–1459. doi: 10.1128/jvi.63.3.1455-1459.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A. K., Huh T. Y., Helleiner C. W. Transcription of satellite DNA in mouse L-cells. Can J Biochem. 1973 May;51(5):529–532. doi: 10.1139/o73-065. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty J. P., Temin H. M. Determination of the rate of base-pair substitution and insertion mutations in retrovirus replication. J Virol. 1988 Aug;62(8):2817–2822. doi: 10.1128/jvi.62.8.2817-2822.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerman M., Temin H. M. Genes with promoters in retrovirus vectors can be independently suppressed by an epigenetic mechanism. Cell. 1984 Dec;39(3 Pt 2):449–467. [PubMed] [Google Scholar]
- Gaubatz J. W., Cutler R. G. Mouse satellite DNA is transcribed in senescent cardiac muscle. J Biol Chem. 1990 Oct 15;265(29):17753–17758. [PubMed] [Google Scholar]
- Harel J., Hanania N., Tapiero H., Harel L. RNA replication by nuclear satellite DNA in different mouse cells. Biochem Biophys Res Commun. 1968 Nov 25;33(4):696–701. doi: 10.1016/0006-291x(68)90352-5. [DOI] [PubMed] [Google Scholar]
- Hu W. S., Temin H. M. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc Natl Acad Sci U S A. 1990 Feb;87(4):1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W. S., Temin H. M. Retroviral recombination and reverse transcription. Science. 1990 Nov 30;250(4985):1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- Hörz W., Altenburger W. Nucleotide sequence of mouse satellite DNA. Nucleic Acids Res. 1981 Feb 11;9(3):683–696. doi: 10.1093/nar/9.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz R. A., Skalka A. M. Generation of diversity in retroviruses. Annu Rev Genet. 1990;24:409–445. doi: 10.1146/annurev.ge.24.120190.002205. [DOI] [PubMed] [Google Scholar]
- Kawai S., Nishizawa M. New procedure for DNA transfection with polycation and dimethyl sulfoxide. Mol Cell Biol. 1984 Jun;4(6):1172–1174. doi: 10.1128/mcb.4.6.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G., Paige C., Gilboa E., Wagner E. F. Expression of a foreign gene in myeloid and lymphoid cells derived from multipotent haematopoietic precursors. Nature. 1985 Nov 14;318(6042):149–154. doi: 10.1038/318149a0. [DOI] [PubMed] [Google Scholar]
- Klarmann G. J., Schauber C. A., Preston B. D. Template-directed pausing of DNA synthesis by HIV-1 reverse transcriptase during polymerization of HIV-1 sequences in vitro. J Biol Chem. 1993 May 5;268(13):9793–9802. [PubMed] [Google Scholar]
- Leider J. M., Palese P., Smith F. I. Determination of the mutation rate of a retrovirus. J Virol. 1988 Sep;62(9):3084–3091. doi: 10.1128/jvi.62.9.3084-3091.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz D., Goff S., Bank A. A safe packaging line for gene transfer: separating viral genes on two different plasmids. J Virol. 1988 Apr;62(4):1120–1124. doi: 10.1128/jvi.62.4.1120-1124.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A. D., Garcia J. V., von Suhr N., Lynch C. M., Wilson C., Eiden M. V. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J Virol. 1991 May;65(5):2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A. D. Human gene therapy comes of age. Nature. 1992 Jun 11;357(6378):455–460. doi: 10.1038/357455a0. [DOI] [PubMed] [Google Scholar]
- Monk R. J., Malik F. G., Stokesberry D., Evans L. H. Direct determination of the point mutation rate of a murine retrovirus. J Virol. 1992 Jun;66(6):3683–3689. doi: 10.1128/jvi.66.6.3683-3689.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J. E., Goff S. P. Construction and analysis of deletion mutations in the U5 region of Moloney murine leukemia virus: effects on RNA packaging and reverse transcription. J Virol. 1989 Jan;63(1):319–327. doi: 10.1128/jvi.63.1.319-327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer C. A., Pogue-Geile K., Guntaka R., Staskus K. A., Faras A. J. Involvement of directly repeated sequences in the generation of deletions of the avian sarcoma virus src gene. J Virol. 1983 Aug;47(2):380–382. doi: 10.1128/jvi.47.2.380-382.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak V. K., Temin H. M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6024–6028. doi: 10.1073/pnas.87.16.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak V. K., Temin H. M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6019–6023. doi: 10.1073/pnas.87.16.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peliska J. A., Benkovic S. J. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992 Nov 13;258(5085):1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- Pulsinelli G. A., Temin H. M. Characterization of large deletions occurring during a single round of retrovirus vector replication: novel deletion mechanism involving errors in strand transfer. J Virol. 1991 Sep;65(9):4786–4797. doi: 10.1128/jvi.65.9.4786-4797.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radic M. Z., Lundgren K., Hamkalo B. A. Curvature of mouse satellite DNA and condensation of heterochromatin. Cell. 1987 Sep 25;50(7):1101–1108. doi: 10.1016/0092-8674(87)90176-0. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Sealy L., Hartley J., Donelson J., Chalkley R., Hutchison N., Hamkalo B. Characterization of a highly repetitive sequence DNA family in rat. J Mol Biol. 1981 Jan 15;145(2):291–318. doi: 10.1016/0022-2836(81)90207-2. [DOI] [PubMed] [Google Scholar]
- Shields A., Witte W. N., Rothenberg E., Baltimore D. High frequency of aberrant expression of Moloney murine leukemia virus in clonal infections. Cell. 1978 Jul;14(3):601–609. doi: 10.1016/0092-8674(78)90245-3. [DOI] [PubMed] [Google Scholar]
- Shimotohno K., Temin H. M. Formation of infectious progeny virus after insertion of herpes simplex thymidine kinase gene into DNA of an avian retrovirus. Cell. 1981 Oct;26(1 Pt 1):67–77. doi: 10.1016/0092-8674(81)90034-9. [DOI] [PubMed] [Google Scholar]
- Shimotohno K., Temin H. M. Spontaneous variation and synthesis in the U3 region of the long terminal repeat of an avian retrovirus. J Virol. 1982 Jan;41(1):163–171. doi: 10.1128/jvi.41.1.163-171.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steck F. T., Rubin H. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. I. Establishment of interference. Virology. 1966 Aug;29(4):628–641. doi: 10.1016/0042-6822(66)90287-x. [DOI] [PubMed] [Google Scholar]
- Stuhlmann H., Berg P. Homologous recombination of copackaged retrovirus RNAs during reverse transcription. J Virol. 1992 Apr;66(4):2378–2388. doi: 10.1128/jvi.66.4.2378-2388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain A., Coffin J. M. Mechanism of transduction by retroviruses. Science. 1992 Feb 14;255(5046):841–845. doi: 10.1126/science.1371365. [DOI] [PubMed] [Google Scholar]
- Varela-Echavarría A., Garvey N., Preston B. D., Dougherty J. P. Comparison of Moloney murine leukemia virus mutation rate with the fidelity of its reverse transcriptase in vitro. J Biol Chem. 1992 Dec 5;267(34):24681–24688. [PubMed] [Google Scholar]
- Voynow S. L., Coffin J. M. Evolutionary variants of Rous sarcoma virus: large deletion mutants do not result from homologous recombination. J Virol. 1985 Jul;55(1):67–78. doi: 10.1128/jvi.55.1.67-78.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voynow S. L., Coffin J. M. Truncated gag-related proteins are produced by large deletion mutants of Rous sarcoma virus and form virus particles. J Virol. 1985 Jul;55(1):79–85. doi: 10.1128/jvi.55.1.79-85.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Boeke J. D. High-frequency deletion between homologous sequences during retrotransposition of Ty elements in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8553–8557. doi: 10.1073/pnas.84.23.8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Temin H. M. 3' junctions of oncogene-virus sequences and the mechanisms for formation of highly oncogenic retroviruses. J Virol. 1993 Apr;67(4):1747–1751. doi: 10.1128/jvi.67.4.1747-1751.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]