Abstract
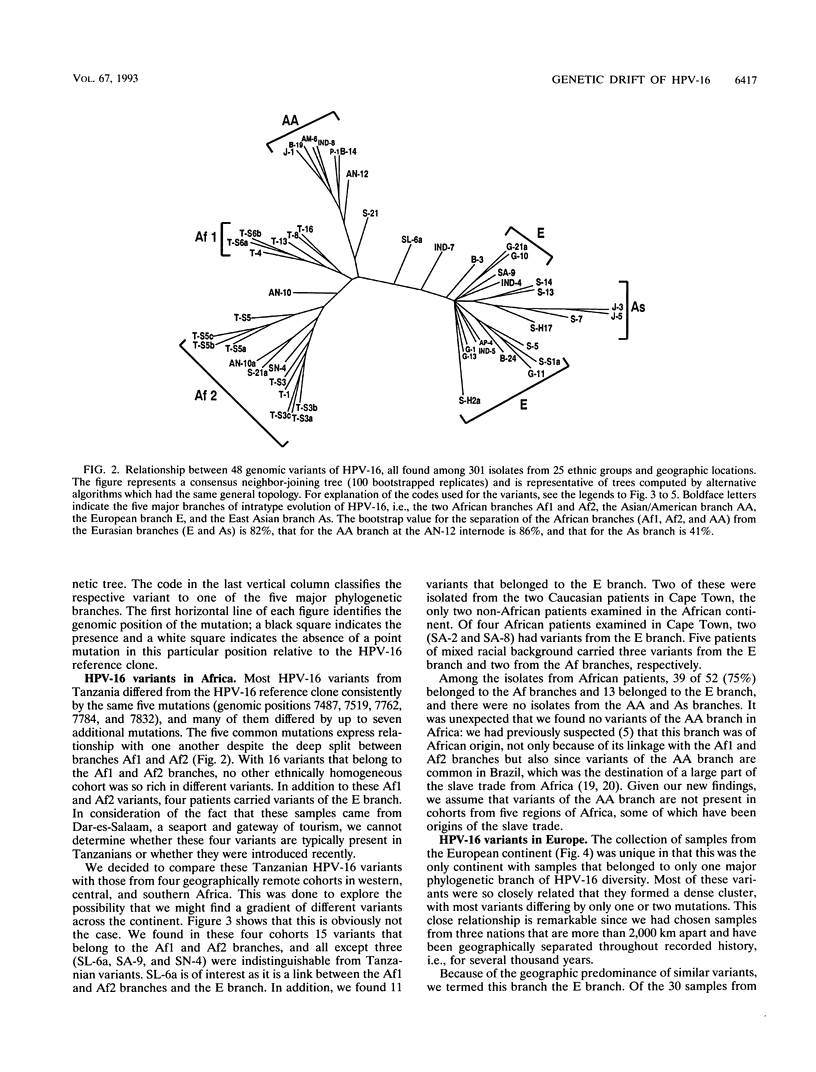
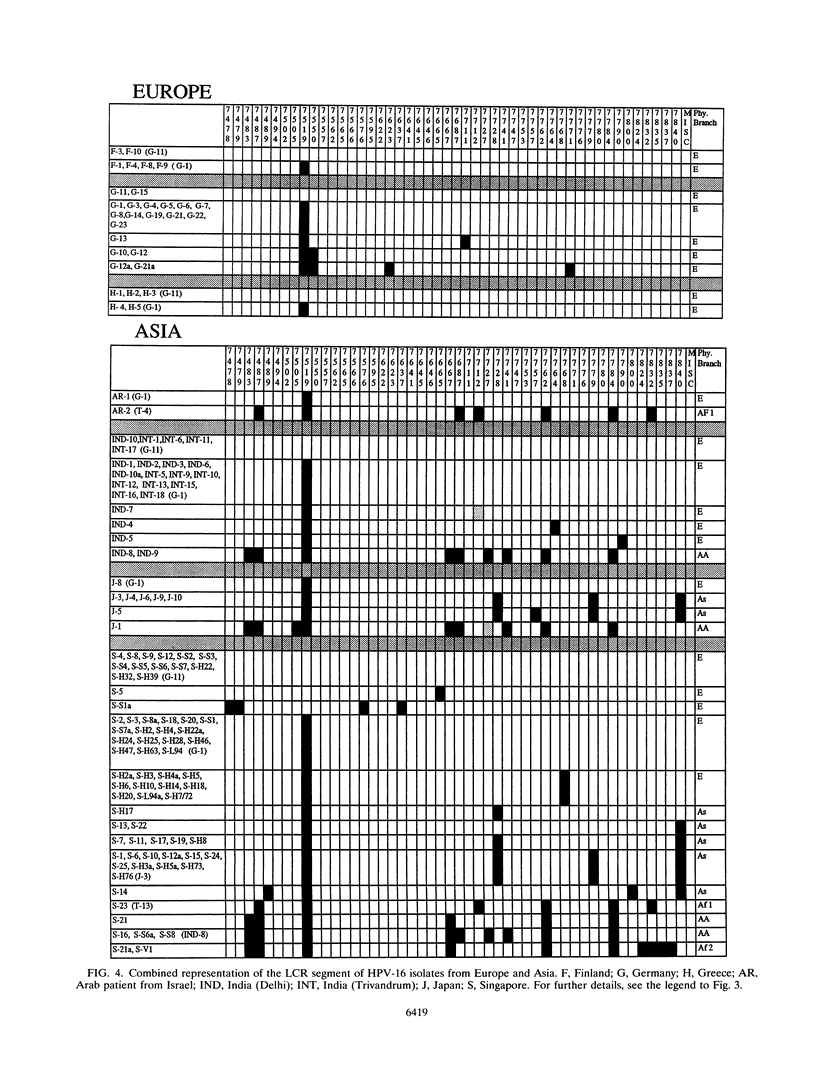
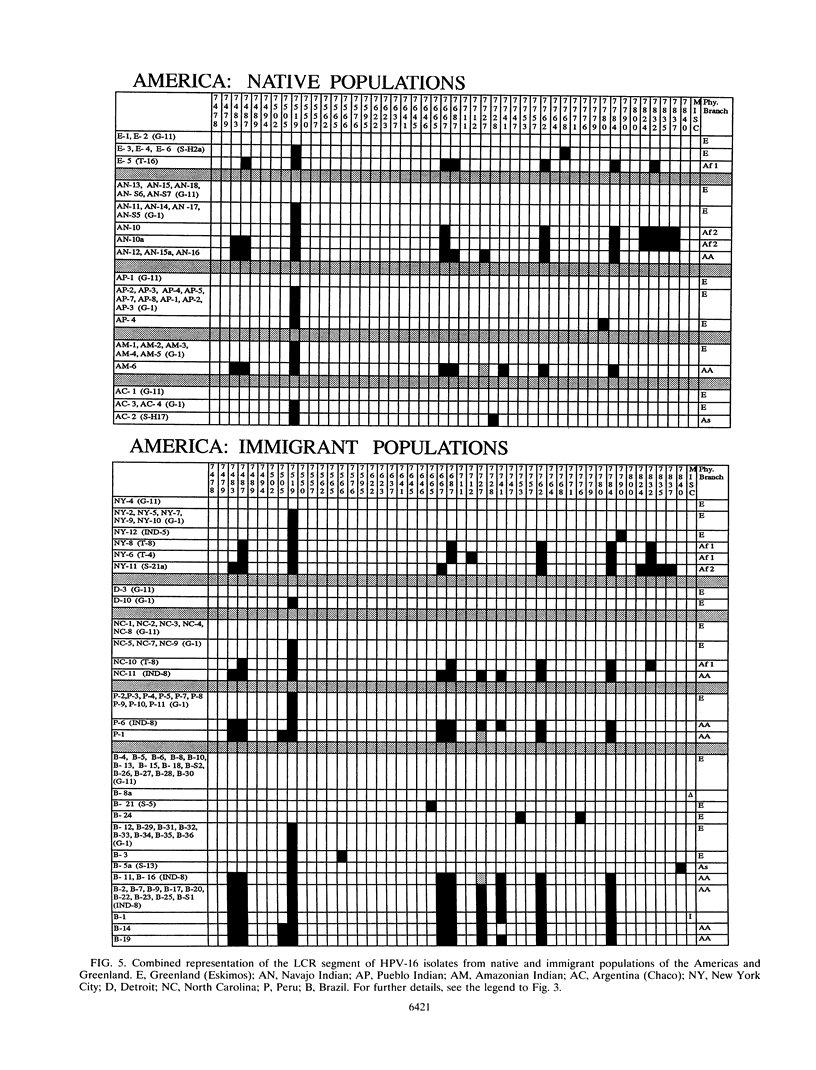
We have investigated the diversity of a hypervariable segment of the human papillomavirus type 16 (HPV-16) genome among 301 virus isolates that were collected from 25 different ethnic groups and geographic locations. Altogether, we distinguished 48 different variants that had diversified from one another along five phylogenetic branches. Variants from two of these branches were nearly completely confined to Africa. Variants from a third branch were the only variants identified in Europeans but occurred at lower frequency in all other ethnic groups. A fourth branch was specific for Japanese and Chinese isolates. A small fraction of all isolates from Asia and from indigenous as well as immigrant populations in the Americas formed a fifth branch. Important patterns of HPV-16 phylogeny suggested coevolution of the virus with people of the three major human races, namely, Africans, Caucasians, and East Asians. But several minor patterns are indicative of smaller bottlenecks of viral evolution and spread, which may correlate with the migration of ethnic groups in prehistoric times. The colonization of the Americas by Europeans and Africans is reflected in the composition of their HPV-16 variants. We discuss arguments that today's HPV-16 genomes represent a degree of diversity that evolved over a large time span, probably exceeding 200,000 years, from a precursor genome that may have originated in Africa. The identification of molecular variants is a powerful epidemiological and phylogenetic tool for revealing the ancient spread of papillomaviruses, whose trace through the world has not yet been completely lost.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cavalli-Sforza L. L., Menozzi P., Piazza A. Demic expansions and human evolution. Science. 1993 Jan 29;259(5095):639–646. doi: 10.1126/science.8430313. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L., Minch E., Mountain J. L. Coevolution of genes and languages revisited. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5620–5624. doi: 10.1073/pnas.89.12.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. Y., Bernard H. U., Ong C. K., Chan S. P., Hofmann B., Delius H. Phylogenetic analysis of 48 papillomavirus types and 28 subtypes and variants: a showcase for the molecular evolution of DNA viruses. J Virol. 1992 Oct;66(10):5714–5725. doi: 10.1128/jvi.66.10.5714-5725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. Y., Ho L., Ong C. K., Chow V., Drescher B., Dürst M., ter Meulen J., Villa L., Luande J., Mgaya H. N. Molecular variants of human papillomavirus type 16 from four continents suggest ancient pandemic spread of the virus and its coevolution with humankind. J Virol. 1992 Apr;66(4):2057–2066. doi: 10.1128/jvi.66.4.2057-2066.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürst M., Gissmann L., Ikenberg H., zur Hausen H. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3812–3815. doi: 10.1073/pnas.80.12.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschle D., Dürst M., ter Meulen J., Luande J., Eberhardt H. C., Pawlita M., Gissmann L. Geographical dependence of sequence variation in the E7 gene of human papillomavirus type 16. J Gen Virol. 1992 Jul;73(Pt 7):1829–1832. doi: 10.1099/0022-1317-73-7-1829. [DOI] [PubMed] [Google Scholar]
- Gessain A., Boeri E., Yanagihara R., Gallo R. C., Franchini G. Complete nucleotide sequence of a highly divergent human T-cell leukemia (lymphotropic) virus type I (HTLV-I) variant from melanesia: genetic and phylogenetic relationship to HTLV-I strains from other geographical regions. J Virol. 1993 Feb;67(2):1015–1023. doi: 10.1128/jvi.67.2.1015-1023.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessain A., Gallo R. C., Franchini G. Low degree of human T-cell leukemia/lymphoma virus type I genetic drift in vivo as a means of monitoring viral transmission and movement of ancient human populations. J Virol. 1992 Apr;66(4):2288–2295. doi: 10.1128/jvi.66.4.2288-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons A. Geneticists trace the DNA trail of the first Americans. Science. 1993 Jan 15;259(5093):312–313. doi: 10.1126/science.8420001. [DOI] [PubMed] [Google Scholar]
- Ho L., Chan S. Y., Chow V., Chong T., Tay S. K., Villa L. L., Bernard H. U. Sequence variants of human papillomavirus type 16 in clinical samples permit verification and extension of epidemiological studies and construction of a phylogenetic tree. J Clin Microbiol. 1991 Sep;29(9):1765–1772. doi: 10.1128/jcm.29.9.1765-1772.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffecker J. F., Powers W. R., Goebel T. The colonization of beringia and the peopling of the new world. Science. 1993 Jan 1;259(5091):46–53. doi: 10.1126/science.259.5091.46. [DOI] [PubMed] [Google Scholar]
- Icenogle J. P., Sathya P., Miller D. L., Tucker R. A., Rawls W. E. Nucleotide and amino acid sequence variation in the L1 and E7 open reading frames of human papillomavirus type 6 and type 16. Virology. 1991 Sep;184(1):101–107. doi: 10.1016/0042-6822(91)90826-w. [DOI] [PubMed] [Google Scholar]
- Ong C. K., Chan S. Y., Campo M. S., Fujinaga K., Mavromara-Nazos P., Labropoulou V., Pfister H., Tay S. K., ter Meulen J., Villa L. L. Evolution of human papillomavirus type 18: an ancient phylogenetic root in Africa and intratype diversity reflect coevolution with human ethnic groups. J Virol. 1993 Nov;67(11):6424–6431. doi: 10.1128/jvi.67.11.6424-6431.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal R. R., Oden N. L., Thomson B. A. Origins of the Indo-Europeans: genetic evidence. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7669–7673. doi: 10.1073/pnas.89.16.7669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ranst M., Kaplan J. B., Burk R. D. Phylogenetic classification of human papillomaviruses: correlation with clinical manifestations. J Gen Virol. 1992 Oct;73(Pt 10):2653–2660. doi: 10.1099/0022-1317-73-10-2653. [DOI] [PubMed] [Google Scholar]
- Vigilant L., Stoneking M., Harpending H., Hawkes K., Wilson A. C. African populations and the evolution of human mitochondrial DNA. Science. 1991 Sep 27;253(5027):1503–1507. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- zur Hausen H. Viruses in human cancers. Science. 1991 Nov 22;254(5035):1167–1173. doi: 10.1126/science.1659743. [DOI] [PubMed] [Google Scholar]



