Abstract
To investigate the role of the neck domain of kinesin, we used optical trapping nanometry to perform high-resolution measurements of the movements and forces produced by recombinant kinesin fragments in which the neck domains were shortened or replaced by an artificial random coil. Truncated kinesin fragments (K351) that contain a motor domain consisting of ≈340 aa and a short neck domain consisting of ≈11 aa showed fast movement (800 nm/s) and 8-nm steps. Such behavior was similar to that of recombinant fragments containing the full-length neck domain (K411) and to that of native kinesin. Kinesin fragments lacking the short neck domain (K340), however, showed very slow movement (<50 nm/s), as previously reported. Joining an artificial 11-aa sequence that was expected to form a flexible random chain to the motor domain (K340–chain) produced normal fast (≈700 nm/s) and stepwise movement. The results suggest that the neck domain does not act as a rigid lever arm to magnify the structural change at the catalytic domain as has been believed for myosin, but it does act as a flexible joint to guarantee the mobility of the motor domain.
Kinesin is a 2 × 2 heterotetramer that produces plus-end-directed movement along microtubules (1). Each kinesin heavy chain has a three-domain structure consisting of a globular N-terminal domain, a long α-helical stalk, and a globular C-terminal domain that binds to a light chain (1). The sequence of the N-terminal ≈340-aa domain (motor domain) with ATPase and microtubule binding sites is strongly conserved among kinesin family members (1). Recent x-ray crystallography revealed that the structure of the motor domain of human kinesin, ≈7 nm × 4.5 nm × 4.5 nm, has striking similarity to that of the core of the catalytic domain of myosin, despite having virtually no amino acid sequence identity (2). Accordingly, it has been suggested that kinesin and myosin share a similar molecular mechanism (2), in which the neck region extending from the catalytic or motor domain acts as a rigid lever arm to magnify the subnanometer structural change at the catalytic domain into a displacement of several nanometers (2–5). Sedimentation studies and secondary structure predictions on Drosophila kinesin have suggested that the region after residue 338 forms α-helix and that dimerization is induced by a coiled-coil interaction between conserved hydrophobic heptad repeats, in the region from amino acid residue 348 to residue 382 (6, 7). This locates the possible hinge at a position from residue 346 to residue 358 (6, 7).
Recent in vitro motility assays have shown that an N-terminal 340-aa kinesin fragment that mostly lacks the neck domain (K340) cannot move as fast as native kinesin or recombinant truncated fragments with a full-size neck domain (8–10). This result suggests that the neck domain of the kinesin molecule plays an important role in generating movements efficiently. To examine the role of the neck domain, we have used optical trapping nanometry to measure the movements of bead-attached recombinant kinesin fragments moving along a microtubule (11–17). We made four kinds of recombinant fragments of Drosophila kinesin with the different sizes of neck domains (K340, K351, and K411) and with a short neck consisting of an artificial 11-aa sequence that probably forms a flexible chain (K340–chain). To minimize the potential for damage to the fragments during interaction with the surface of a bead, a reactive cysteine was introduced into the C termini of the fragments and the introduced cysteine was biotinylated. The biotinylated fragments attached to the streptavidin-coated bead with the motor domain of the fragments distal from the bead surface (9, 18, 19). The motor domain lacking the neck domain (K340) showed very slow movement as reported (8–10), but K351 and K340–chain could produce normal fast and stepwise movement, similar to that of native kinesin. The results suggest that the neck domain does not act as a rigid lever arm to magnify the structural change at the motor domain but rather acts as a flexible joint to guarantee the mobility of the motor domain. Furthermore, because it is unlikely that the chain artificially introduced into K340 causes the motor domains to dimerize, the interhead coupling specified by the authentic kinesin neck would not be essential for normal movement. A preliminary reports of these results have appeared (19, 20).
MATERIALS AND METHODS
Proteins.
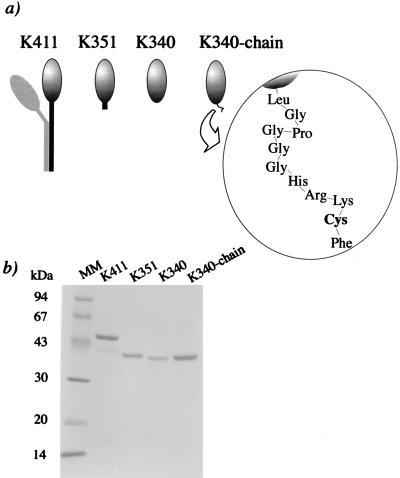
The Drosophila kinesin genes were truncated at amino acid residue 340 (K340), residue 351 (K351), and residue 411 (K411). To biotinylate the C termini, -Lys-Arg-Cys, -Cys, and -Lys-Arg-Cys were introduced at the C termini of K340, K351, and K411, respectively. K340 with the C terminus of -Leu-Gly-Pro-Gly-Gly-Gly-His-Arg-Lys-Cys-Phe (K340–chain) was also made (Fig. 1). These constructs were expressed in Escherichia coli as fusions with glutathione S-transferase (GST) by the method of Stewart et al. (8). GST–kinesin fragments were purified by a glutathione-agarose column and repeated cycles of microtubule binding and dissociation. GST was then removed by proteolysis with thrombin. The cysteine residues at the C termini of kinesin fragments were reacted with maleimide-biotin (18, 19). The oligomeric states of kinesin fragments in solution were determined by 7–20% sucrose gradient centrifugation.
Figure 1.
(a) Illustrations of recombinant truncated kinesin fragments. Each fragment has reactive cysteine residues at the C termini. (b) SDS/PAGE of purified fragments. Approximately 1 μg of each fragment was loaded on a 15% polyacrylamide gel. Lane MM, molecular mass makers whose positions in kilodaltons are indicated to the left. The molecular masses of K411, K351, K340, and K340–chain were consistent with those calculated from the amino acid sequence, 46.2 kDa, 39.2 kDa, 38.0 kDa, and 38.8 kDa, respectively.
Kinesin-Coated Beads.
Carboxylated fluorescent beads 1 μm in diameter were crosslinked with streptavidin via a 6-amino-n-caproic acid linkage by using a carbodiimide kit (Polysciences). Streptavidin-coated beads were suspended in a solution of biotinylated all fragments (0.01–0.1 μg/ml) for 30 min at 25–27°C to allow binding of the kinesin fragments to the beads. Contaminating ATP and unbound kinesin fragments were removed by centrifugation.
The concentration of fragments was chosen so that the fraction of moving bead was 0.1–0.5, when the kinesin-coated beads were positioned to make contact with a microtubule. By using the statistical analysis by Svoboda et al. (11, 12), the number of minimum functional units, single molecules in the case of native kinesin, involved in force generation is almost (>99%) unity at a motive fraction of <0.5.
Motility Assay and Optical Trapping Nanometry.
Motility assay of beads and an apparatus of trapping nanometry were as described previously (17). Taxol-stabilized microtubules visualized by copolymerizing tetramethylrhodamine-labeled tubulin and nonfluorescent tubulin in a molar ratio of 1:20 were bound tightly to the coverslip. By using an optical trap with a trap stiffness of 0.02–0.07 pN/nm, a kinesin-coated bead was placed in contact with a microtubule. The displacement of the bead was measured with a quadrant photodiode detector with nanometer accuracy (17, 21). The displacement by kinesin fragments was determined from the bead displacement as (bead displacement) × [(Kt + Kp)/Kp], where Kt and Kp are, respectively, the stiffness of the optical trap and the stiffness of bead-to-glass linkage, which is the series linkages of a bead, kinesin fragments, a microtubule, and the surface of coverslip (17, 22). Kp was monitored simultaneously by applying sinusoidal displacement changes with the amplitude of 5 nm and the frequency of 100 Hz to the bead (17). Data were recorded at a sampling rate of 200 s−1 after passing through a low-pass filter with a cut-off frequency of 50 Hz. Experiments were performed in a buffer solution containing 80 mM Pipes, 1 mM EGTA, 1 mM ATP, and 2 mM MgCl2, pH 6.8 at 25–27°C.
RESULTS AND DISCUSSION
The Short Neck Domain Is Important for Normal Movement.
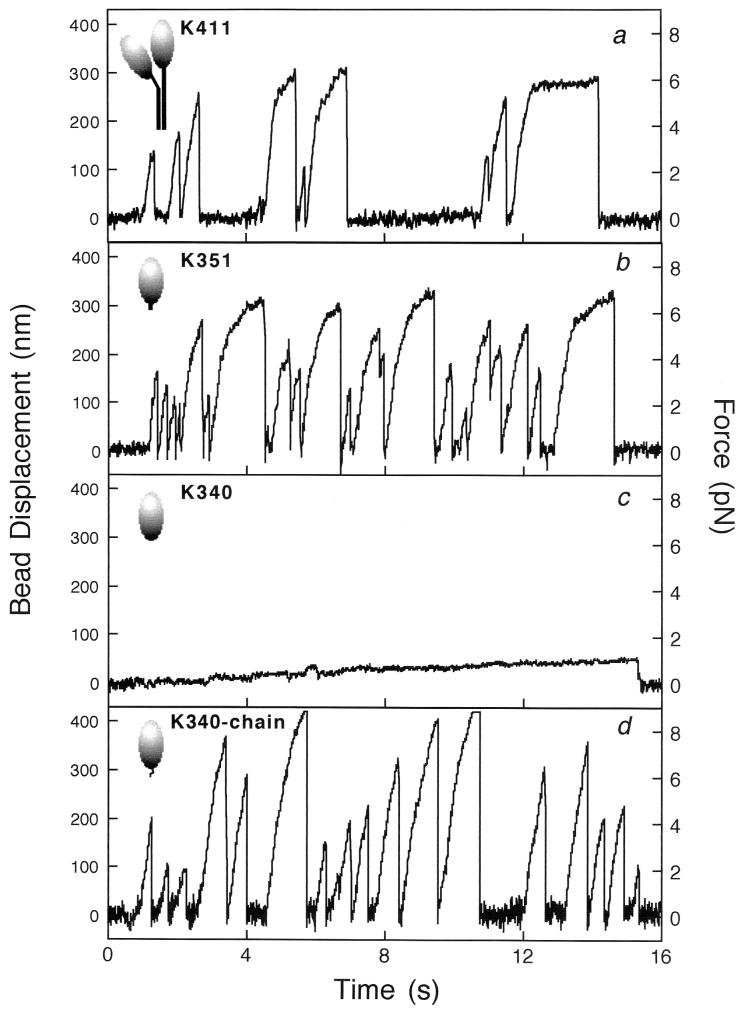
Fig. 2 shows the bead displacement and the force exerted by K411, K351, K340, and K340–chain. The rise times and stall forces for K411 and K351 were quite similar to those of single native kinesin molecules (11, 23, 24). The movement by K340 was very slow, consistent with previous work (8–10). When an 11-aa artificial sequence was fused to the C terminus of K340, the velocity and the force were recovered.
Figure 2.
Time courses of displacement and force transients produced by truncated kinesin fragments, K411 (a), K351 (b), K340 (c), and K340–chain (d). The C termini of recombinant fragments were biotinylated to specifically bind to streptavidin-coated beads. By using an optical trap, a kinesin-coated bead was positioned to make contact with a microtubule adsorbed on a glass surface, and its displacements were detected by a quadrant photodiode detector with nanometer accuracy. The forces were obtained as (bead displacement) × (trapping stiffness).
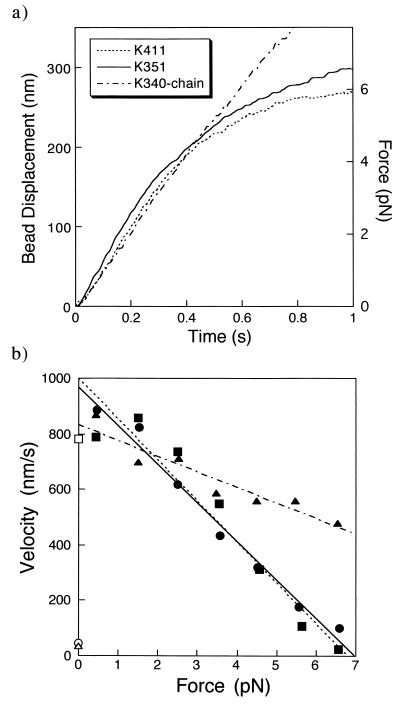
Fig. 3a shows the averaged time courses of 30 runs by K411, K351, and K340–chain. Fig. 3b shows the relationships between the forces and velocities during the transients. The velocities were obtained from the slopes of the averaged time courses of the transients shown in Fig. 3a. As the displacements of kinesins are attenuated due to the stiffness of the bead-to-glass linkage, the bead displacements were corrected as described in Materials and Methods. The velocities of K411, K351, and K340–chain were all approximately 800 nm/s at low forces of <2 pN. The maximum forces of K411 and K351 were about 7 pN (Fig. 3). This behavior was similar to that of native kinesins (11, 17, 23, 24). It is not known how many molecules were involved in generating the movements. But, as the velocity at low load is independent of the number of molecules involved (10, 25), the results show that K411, K351, and K340–chain can produce normal fast movement that is qualitatively similar to that of native kinesins.
Figure 3.
Force–velocity relationships. (a) Time courses of the displacement transients averaged for 30 events by K411 (dotted line), K351 (solid line), and K340–chain (broken line). (b) Force–velocity relationships of K411 (▪), K351 (•), and K340–chain (▴). The velocities were calculated from the slopes of the displacement transients averaged for every 1 pN of force, taking into account of the attenuation, [(Kt + Kp)/Kp], due to the compliance of bead-to-glass linkage. Dotted, solid, and broken lines indicate the best fits to the data for K411, K351, and K340–chain, respectively. The gliding velocities of microtubules by K411 (□), K351 (○), and K340 (▵) attached to the glass surface by a biotin-streptavidin system are also shown.
It was reported (10) that in a microtubule gliding assay, K350 and K365 bound to a coverslip move a microtubule very slowly, at about one-tenth of the velocity of native kinesin. We also found in the gliding assay that the velocity of a microtubule produced by K351 was very low (46 ± 2 nm/s; mean ± SEM, n = 42), about one-twentith of the maximum velocity of K351 in the present bead assay. The difference of velocities between the gliding and bead assays would not result from the linkage between the kinesin fragments and the surface, because both assays used the same biotin-streptavidin linkage. In the gliding assay, many K351 fragments would interact with a microtubule. All K351 molecules could not correctly orient relative to the microtubule axis because K351 molecules were randomly oriented on the surface. Incorrectly oriented K351 molecules could not produce normal fast movement and would provide friction to the fast movement by correctly oriented molecules as observed for myosin (26, 27). On the other hand, in the bead assay, a bead captured in an optical trap can freely rotate (28) and the number of molecules that can interact with the microtubule would be smaller, probably one or two, so a larger fraction of molecules could easily change orientation to fit the heads to the microtubule. K411 moved at similar speeds in surface and bead assays, suggesting that the long neck domain tailing the head guarantees sufficient mobility of a head, independent of whether the molecule is bound to the bead or the surface.
In conclusion, the whole neck domain is not essential for generating normal movement and force, but the short neck from residue 341 to residue 351 is important for generating normal fast movement and normal force.
Role of the Short Neck Domain.
To examine whether the short neck domain from residue 341 to residue 351 acts as a rigid lever arm or as a flexible joint between the head and the surface of a substrate, the 11 aa at the C terminus of K351 were replaced by an 11-aa artificial sequence, -Leu-Gly-Pro-Gly-Gly-Gly-His-Arg-Lys-Cys-Phe (Fig. 1). The sequence is predicted not to form a rigid secondary structure by the programs by Solovyev et al. (29) (http://dot.imgen.bcm.tmc.edu:9331/pssprediction/pssp.html), which typically show more than 70% prediction accuracy. This replacement changed the maximum velocity only very slightly (Fig. 3b). The results suggest that the neck domain does not act as a rigid lever arm to magnify the structural change in the motor domain in proportion to the length of the neck domain as proposed for myosin (3, 4, 30), but rather, it may act as a flexible joint between the head and the substrate to guarantee the mobility of the head.
Role of the Long Neck Region.
Motilities of K411- and K351-coated beads along microtubules were quite similar to each other in the trap. When kinesin-coated beads that had moved in the trap were released from trapping by turning off a trap laser, most of the K411-coated beads (85%, 17 of 20 beads) moved processively for more than 1 μm, whereas few of the K351-coated beads (10%, 2 of 20 beads) moved processively. Thus, the long neck domain is important for generating processive movement under free diffusion without trapping, probably by dimerizing the motor domains (see below). To our knowledge, this is the first report that a monomeric kinesin construct cannot move processively along microtubules even when it can move as rapidly as the native kinesin.
Is the Two-Headed Structure Essential for Generating Normal Movement and Force?
A sedimentation study showed that K340 and K351 were monomeric, i.e., one-headed in solution, whereas K411 forms dimers (data not shown), in good agreement with previous work (5–10). As a streptavidin molecule has four binding sites for biotin, it is possible that K351 molecules or K340–chains that bind to adjacent sites on a streptavidin molecule would move cooperatively just as well as two heads of the native kinesin. Nevertheless, we emphasize that these two single-headed fragments could not make strong contact within the region from residue 340 to residue 351, even if the fragments bind to the same streptavidin, because that region has no heptad repeats of conserved hydrophobic residues to form a coiled-coil interaction (6). Furthermore, the interval of adjacent biotin binding sites of a streptavidin, >1.5 nm (31), is similar to or less than the length (<1.7 nm) of that region expected if it forms α-helix. We conclude that the interhead coupling specified by the authentic kinesin neck is not essential for normal fast movement.
When the beads were released from the laser trap, K351 could not move the beads a long distance as stably as K411 and a native kinesin. Therefore, the authentic two-headed structure is important for generating the movement stably, probably in a hand-over-hand fashion (5, 6, 9).
Minimum Number of Molecules Necessary for Producing Normal Movement.
The density of kinesin fragments bound to a bead was chosen so that the fraction of moving bead was 0.1–0.5, where the number of functional units involved in force generation is almost (>99%) one. As K411 is a two-headed structure and produced similar force to a single native kinesin molecule, the number of K411s involved in the force generation would be one. K351 also generated a force–velocity curve similar to that of K411 and to native kinesin. Although the possibility cannot be excluded that a single one-headed K351 might move and generate force as well as a native one, it is more likely that two K351 molecules attached to a bead operated cooperatively to produce movement. For K340–chain, more molecules would be involved, because larger forces were observed. If the stall force per head is constant, as the slope of force vs. velocity curve is approximately half that of two-headed constructs (Fig. 3b), the number of molecules involved would be approximately four.
Stepwise Movements.
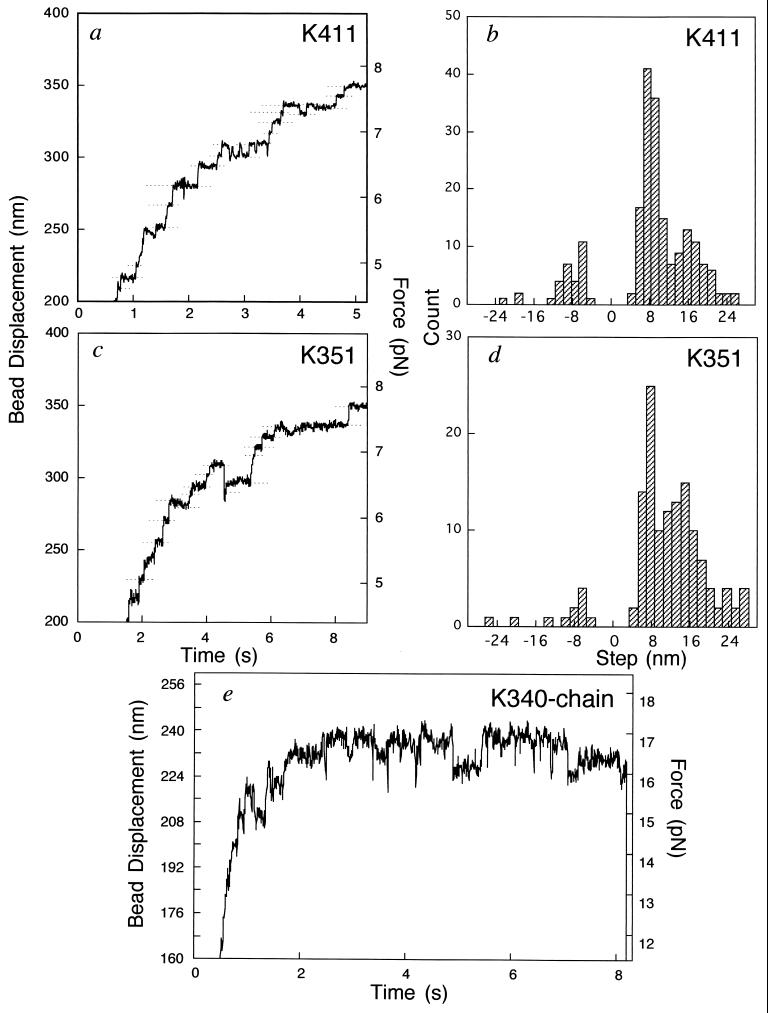
Fig. 4 a, c, and e shows expanded time courses of bead displacements for K411, K351, and K340–chain, respectively. K411, K351, and K340–chain exhibited forward and occasionally backward stepwise displacements. Fig. 4 b and d shows the histograms of the step size for K411 and K351, respectively. In both cases, the step sizes were predominantly 8 nm, which coincides with the periodicity of the α/β-tubulin dimmer in a microtubule. Larger steps of ≈16 and ≈24 nm are also seen but are due probably to multiple 8-nm steps occurring within the time resolution (12, 17). Such stepwise movements closely resemble those observed for single native kinesin molecules (12, 14, 15). K340–chain sometimes also showed stepwise motions, with amplitudes of near 8 nm or multiples of 8 nm, but they were not as clear as those of K411 and K351 (Fig. 4e). This is probably because more heads, probably four heads, are involved, so that steps by individual heads would be damped by the compliance due to other heads. Thus, the elementary process of force generation of the truncated kinesin fragments of K411, K351, and K340–chain would be essentially the same as that of native kinesins.
Figure 4.
Stepwise movements. (a and c) Expanded time courses of K411-bead and K351-bead displacements at trapping stiffness of 0.022 pN/nm. Horizontal broken lines show the positions averaged in the regions where a bead paused within a standard deviation of 2 nm for longer than 30 ms (b and d). Histograms of the step sizes of K411 and K351, respectively. The step sizes of kinesin fragments shown in b and d were obtained from the differences between adjacent broken lines after corrected for the compliance due to the bead-to-glass linkage. The attenuation due to the stiffness of the bead-to-glass linkage [(Kt + Kp)/Kp] was 1.1–1.3 in the region in which we counted the events. (e) Expanded time course of one K340–chain bead at a trapping stiffness 0.071 pN/nm.
CONCLUSIONS
The results suggest that the neck domain does not act as a rigid lever arm to magnify the structural change at the motor domain but as a flexible joint to guarantee the mobility of the motor domain. Furthermore, because it is unlikely that the chain artificially introduced into K340 dimerizes the motor domains, the interhead coupling specified by the authentic kinesin neck would not be essential for normal fast movement.
Acknowledgments
We thank Dr. L. S. B. Goldstein for kindly providing the clone of Drosophila kinesin cDNA, Dr. V. V. Solovyev for offering the programs of secondary structure prediction, and Dr. R. A. Cross for critical reading of this manuscript. This work was supported, in part, by Japan Society for the Promotion of Science Research Fellowships for Young Scientists.
References
- 1.Bloom G, Endow S. Motor Proteins. Vol. 1. London: Academic; 1994. [Google Scholar]
- 2.Kull F J, Sablin E P, Lau R, Fletterick R J, Vale R D. Nature (London) 1996;380:550–555. doi: 10.1038/380550a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rayment I, Holden H M, Whittaker M, Yohn C B, Lorentz M, Holmes K C, Milligan R A. Science. 1993;261:58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- 4.Spudich J A. Nature (London) 1994;372:515–518. doi: 10.1038/372515a0. [DOI] [PubMed] [Google Scholar]
- 5.Hirose K, Lockhart A, Cross R A, Amos L A. Nature (London) 1995;376:277–279. doi: 10.1038/376277a0. [DOI] [PubMed] [Google Scholar]
- 6.Huang T G, Suhan J, Hackney D D. J Biol Chem. 1994;269:16502–16507. [PubMed] [Google Scholar]
- 7.Correia J J, Gilbert S P, Moyer M L, Johnson K A. Biochemistry. 1995;34:4898–4907. doi: 10.1021/bi00014a047. [DOI] [PubMed] [Google Scholar]
- 8.Stewart R J, Thaler J P, Goldstein L S B. Proc Natl Acad Sci USA. 1993;90:5209–5213. doi: 10.1073/pnas.90.11.5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berliner E, Young E C, Anderson K, Mahtani H K, Gelles J. Nature (London) 1995;373:718–721. doi: 10.1038/373718a0. [DOI] [PubMed] [Google Scholar]
- 10.Vale R D, Funatsu T, Pierce D W, Romberg L, Harada Y, Yanagida T. Nature (London) 1996;380:451–453. doi: 10.1038/380451a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Svoboda K, Block S M. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 12.Svoboda K, Schmidt C F, Schnapp B J, Block S M. Nature (London) 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 13.Kuo S C, Sheetz M P. Science. 1993;260:232–234. doi: 10.1126/science.8469975. [DOI] [PubMed] [Google Scholar]
- 14.Coppin C M, Finer J T, Spudich J A, Vale R D. Proc Natl Acad Sci USA. 1996;93:1913–1917. doi: 10.1073/pnas.93.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kojima H, Muto E, Higuchi H, Yanagida T. Biophys J. 1996;70:36. doi: 10.1016/S0006-3495(97)78231-6. (abstr.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higuchi H, Inoue Y, Muto E, Yanagida T. Jpn J Physiol. 1995;45:89s. (abstr.). [Google Scholar]
- 17.Higuchi, H., Muto, E., Inoue, Y. & Yanagida, T. (1997) Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 18.Itakura S, Yamakawa H, Toyoshima Y Y, Ishijima A, Kojima H, Harada Y, Yanagida T, Wakabayashi T, Sutoh K. Biochem Biophys Res Commun. 1993;196:1504–1510. doi: 10.1006/bbrc.1993.2422. [DOI] [PubMed] [Google Scholar]
- 19.Inoue Y, Toyoshima Y Y, Higuchi H, Yanagida T. Biophys J. 1996;70:36. (abstr.). [Google Scholar]
- 20.Inoue Y, Sayuri M, Atsuko H I, Toyoshima Y Y, Higuchi H, Yanagida T. Biophys J. 1997;72:62. (abstr.). [Google Scholar]
- 21.Simmons R M, Finer J T, Chu S, Spudich J A. Biophys J. 1996;70:1813–1822. doi: 10.1016/S0006-3495(96)79746-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishijima A, Doi T, Sakurada K, Yanagida T. Nature (London) 1991;352:301–306. doi: 10.1038/352301a0. [DOI] [PubMed] [Google Scholar]
- 23.Meyhöfer E, Howard J. Proc Natl Acad Sci USA. 1995;92:574–578. doi: 10.1073/pnas.92.2.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muto E, Kojima H, Yanagida T. Jpn J Physiol. 1995;45:89s. (abstr.). [Google Scholar]
- 25.Howard J, Hudspeth A J, Vale R D. Nature (London) 1989;342:154–158. doi: 10.1038/342154a0. [DOI] [PubMed] [Google Scholar]
- 26.Ishijima A, Kojima H, Higuchi H, Harada Y, Funatsu T, Yanagida T. Biophys J. 1996;70:383–400. doi: 10.1016/S0006-3495(96)79582-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sellers J R, Kachar B. Science. 1990;249:406–408. doi: 10.1126/science.2377894. [DOI] [PubMed] [Google Scholar]
- 28.Tsuda Y, Yasutake H, Ishijima A, Yanagida T. Proc Natl Acad Sci USA. 1996;93:12937–12942. doi: 10.1073/pnas.93.23.12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salamov A A, Solovyev V V. J Mol Biol. 1995;247:11–15. doi: 10.1006/jmbi.1994.0116. [DOI] [PubMed] [Google Scholar]
- 30.Uyeda T Q P, Abramson P D, Spudich J A. Proc Natl Acad Sci USA. 1996;93:4459–4464. doi: 10.1073/pnas.93.9.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber P C, Ohlendorf D H, Wendoloski J J, Salemme F R. Science. 1989;243:85–88. doi: 10.1126/science.2911722. [DOI] [PubMed] [Google Scholar]