Abstract
Structural protein 4.1 was first characterized as an important 80-kDa protein in the mature red cell membrane skeleton. It is now known to be a member of a family of protein isoforms detected at diverse intracellular sites in many nucleated mammalian cells. We recently reported that protein 4.1 isoforms are present at interphase in nuclear matrix and are rearranged during the cell cycle. Here we report that protein 4.1 epitopes are present in centrosomes of human and murine cells and are detected by using affinity-purified antibodies specific for 80-kDa red cell 4.1 and for 4.1 peptides. Immunofluorescence, by both conventional and confocal microscopy, showed that protein 4.1 epitopes localized in the pericentriolar region. Protein 4.1 epitopes remained in centrosomes after extraction of cells with detergent, salt, and DNase. Higher resolution electron microscopy of detergent-extracted cell whole mounts showed centrosomal protein 4.1 epitopes distributed along centriolar cylinders and on pericentriolar fibers, at least some of which constitute the filamentous network surrounding each centriole. Double-label electron microscopy showed that protein 4.1 epitopes were predominately localized in regions also occupied by epitopes for centrosome-specific autoimmune serum 5051 but were not found on microtubules. Our results suggest that protein 4.1 is an integral component of centrosome structure, in which it may play an important role in centrosome function during cell division and organization of cellular architecture.
Individual isoforms of a structural protein may have specialized molecular interactions at distinct subcellular locations. We are studying structural protein 4.1, first identified as an 80-kDa membrane skeleton protein of human red blood cells (1, 2). This initial picture has subsequently become increasingly complex. Protein 4.1 is now known to be a member of a large diverse family of proteins generated by extensive alternative RNA splicing with variable usage of translation initiation sites and several types of posttranslational modifications (refs. 3–7; reviewed in ref. 8). Many nucleated animal cells contain 4.1-immunoreactive protein species ranging in size from 30 to 210 kDa (9–11). Protein 4.1 epitopes have been identified in cytoplasmic, nuclear, and perinuclear regions and in the Golgi apparatus (12–19). However, the functions of 4.1 isoforms, other than in mature red cells, have yet to be defined.
In previous investigations, we observed that 4.1 epitopes are distributed throughout the interior of the nucleus and are associated with electron dense nuclear substructures by electron microscopy. Several protein 4.1 isoforms fractionate with nuclear matrix proteins (19). This result suggested that protein 4.1 isoforms may contribute significantly to nuclear architecture and function. Interestingly, 4.1 epitopes also undergo spatial rearrangements during the cell cycle, including localization to the mitotic spindle during mitosis and to the region of centrosomes at interphase. This result suggested that some protein 4.1 isoforms may have roles in the modulation of cytoskeletal organization and cell division by centrosomes.
Although the centrosome has long been identified morphologically as a microtubule organizing center, its substructure and molecular components are not completely characterized. In most animal cells, the centrosome lies close to the nucleus and consists of a pair of centrioles surrounded by a dense fibrogranular area called the pericentriolar material (PCM) (20). Centrioles are interconnected cylindrical structures composed of nine sets of triplet microtubules (α- and β-tubulin polymers) and have morphological similarities to the basal body of sperm and cilia. In the PCM there are appendages extending from centrioles, filaments, and aggregated material called “satellites.” Microtubule growth initiates in the PCM at sites containing a specialized tubulin, γ-tubulin (21, 22). After duplicating, centrosomes migrate to opposite sides of the nucleus and serve as spindle poles during mitosis (reviewed in refs. 23–25).
We report here that protein 4.1 is a component of mammalian centrosome structure. Using resinless whole mount electron microscopy, we obtained detailed information on localization of protein 4.1 within this organelle.
METHODS
Materials.
WI38 cells (CCL 75), CaSki cells (CRL 1550), and 3T3 cells (CCL 92) were obtained from the American Type Culture Collection (Rockville, MD). HCA cells were a gift of J. Campisi (Lawrence Berkeley National Laboratory). NHEK 4138 cells were from Clonetics (Walkersville, MD). Anti-β-tubulin and gold-bead-conjugated secondary antibodies were from Amersham. Fluorescein isothiocynante-conjugated goat anti-rabbit IgG was from Molecular Probes. Tetramethyl-rhodamine-conjugated goat anti-mouse IgG was from Sigma. Autoimmune 5051 serum was the gift of S. Doxsey (University of Massachusetts Medical Center, Worcester, MA).
Protein 4.1 Antibodies.
Antibodies against protein 4.1 and its peptide domains have been described (15, 19). All IgGs were affinity-purified against the immunizing antigen. Briefly, anti-RBC 80-kDa 4.1 was raised against purified human red cell 80-kDa protein 4.1, anti-10–1 against a peptide in exon 16 (the spectrin-actin binding domain), anti-24–2 against a peptide in exon 19, and anti-24–3 against 21 amino acids at the C terminus in exon 21. All antibodies detected red cell 80-kDa protein 4.1 by Western blot analysis and membrane-associated protein 4.1 in intact human red cells by immunofluorescence microscopy.
Indirect Immunofluorescence.
Cells were cultured, fixed in either methanol or formaldehyde/Triton X-100, and immunostained as described (19). Controls using equivalent amounts of nonimmune IgG or without primary antibody were included in each experiment. Samples stained with 4′,6-diamino-2-phenylindole and mounted on slides using Vectorshield (Vector Laboratories) were imaged by using an epifluorescence microscope with a 63× objective (1.4 numerical aperture) oil immersion lens. A Laser Scanning Microscope 410 (Zeiss) was used for confocal microscopy.
Electron Microscopy of Embedment-Free Cell Whole Mounts.
Human fibroblasts grown on parlodion-carbon-coated sterile nickel grids (Ted Pella, Redding CA) were extracted in cytoskeletal buffer, fixed, and immunostained as described (26, 27). Samples were fixed in 2% glutaraldehyde in 0.1 M sodium cacodylate, pH 7.4, washed in cacodylate buffer, dehydrated in ethanol, and critical point dried. The sample grids were thinly coated with carbon and viewed by using a JEOL 1200 EX electron microscope. When imaged for stereo, the total tilt angle was 10°.
RESULTS
Localization of Protein 4.1 in Centrosomes.
Antibodies directed against 80-kDa red cell protein 4.1 and against several 4.1 peptide domains were used to stain CaSki cells for immunofluorescence. These antibodies labeled bright punctate objects close to the nucleus in interphase cells and spindle poles in mitotic cells (Fig. 1 A and B). In addition to the punctate staining, there was weaker staining in the nucleus and sometimes in the cytoplasm (19). Similar punctate perinuclear staining was observed in human fibroblasts (WI38 and HCA cells), human keratinocytes (NHEK 4183 cells), and murine 3T3 fibroblasts (data not shown). The 4.1 epitopes were firmly bound in the centrosomal region because they survived the vigorous extraction with detergent and high salt used to prepare nuclear matrix-intermediate scaffold (Fig. 1C) (26). To confirm 4.1 localization at centrosomes, cells were simultaneously probed with centrosome-specific 5051 autoimmune serum and with 4.1 antibodies. The punctate immunofluorescent labeling by protein 4.1 antibody was coincident with 5051 immunofluorescent signals (Fig. 1D).
Figure 1.
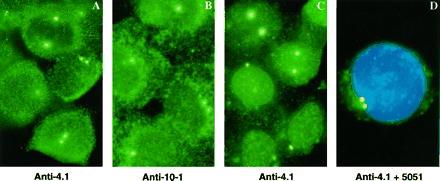
Immunofluorescent localization of protein 4.1 epitopes in centrosomes. CaSki cells were extracted with detergent, fixed with paraformaldehyde, and immunostained. (A) Antibody directed against 80-kDa protein 4.1 (green). (B) Antibody against protein 4.1 spectrin-actin binding domain (anti-10–1; green). Similar perinuclear punctate staining was observed with 4.1 antibodies 24–2 and 24–3. (C) Cells were extracted with detergent, salt, and DNase (19, 26) before fixation and staining with anti-4.1 (green). (D) Staining with anti-4.1 (green), 5051 serum (red), and 4′,6-diamino-2-phenylindole (blue). The yellow perinuclear dots indicate colocalization of red and green signals.
Distribution of Protein 4.1 Epitopes Relative to Centrioles by Immunofluorescence.
Centrioles were located by immunostaining for a major constituent, tubulin. However, before the centriolar tubulin could be seen, the much larger amount of tubulin in microtubules must be removed. Cells were extracted at 4°C in buffers containing calcium to dissociate the dense microtubular network which converges at centrosomes. Samples were then immunolabeled with antibodies against β-tubulin and 80-kDa protein 4.1 (Fig. 2). Confocal microscopy showed centriolar cylinders labeled by anti-β-tubulin (Fig. 2, red) in various longitudinal and perpendicular positions relative to the coverslip. Protein 4.1 epitopes (Fig. 2, green) surrounded the centriolar cylinders. In favorable end-on orientations, protein 4.1 epitopes appeared to cluster on discrete extensions radiating into the PCM and on structures apparently connecting the centrioles.
Figure 2.
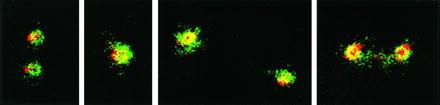
Double-label immunofluorescent microscopy of centrosomes in Caski cells. CaSki cells were extracted with detergent and fixed under conditions that destabilized microtubules. Samples were probed simultaneously with antibodies against β-tubulin (red) to image centriole cylinders and against 80-kDa 4.1 (green). Colocalized red and green signals appear as yellow. Representative centrioles are shown in different orientations.
Localization of Protein 4.1 Epitopes at Centrosomes by Electron Microscopy of Extracted, Unembedded Cell Whole Mounts.
The higher resolution of electron microscopy affords more detailed images of protein 4.1 epitopes and associated centrosome structures. In this report, we used electron microscopy of detergent-extracted cell whole mounts to localize protein 4.1 epitopes in centrosomes of human diploid fibroblasts. Gold-bead-conjugated secondary antibodies against anti-4.1 densely labeled a region adjacent to the nucleus easily identifiable, by its centrioles, as the centrosome (Fig. 3). Gold beads decorated centrioles (seen in longitudinal and end-on orientations) and fibrous structures extending into the PCM. Often 4.1 epitopes were distributed on centriolar cylinders more heavily at one end than at the other (Fig. 3). The sparsely labeled area may correspond to the ends of centrioles that we observed protruding from 4.1-stained areas in immunofluorescence images (Fig. 2).
Figure 3.

Electron micrographs of immunostained, detergent-extracted whole mount human fibroblasts. Protein 4.1 epitopes were detected by using 10 nm gold beads. (Left) A field showing the nucleus (N) and a centrosome in the perinuclear area. The two dense bodies (arrows) in the centrosome are a pair of centrioles. (Right) Two other images of centrosomes heavily decorated with gold beads on centrioles and fibrous structures projecting into the PCM. (Bar = 200 nm.)
Double-Label Immunoelectron Microscopy.
Resinless electron micrographs can show colocalization of antigens in far greater detail compared with optical microscopy. Human serum 5051 detects pericentrin, a 220-kDa antigen, identified as a PCM component (28). Immunofluorescence showed 4.1 epitopes coincident with 5051 epitopes (Fig. 1D). Fig. 4 shows a more precise localization of 4.1 and 5051 epitopes by immunogold labeling of cell whole mounts with different sized gold beads for the two antibodies. The extracted whole mounts were prepared both with microtubules intact (Fig. 4A) and depolymerized (Fig. 4B). For intact microtubules, cells were preincubated for 10 min with 5 μg/ml taxol at 37°C and then extracted and processed at room temperature in buffers containing taxol (Fig. 4A). For depolymerized microtubules, taxol was omitted, and the cells were extracted at 4°C with buffers containing 2 mM calcium (Fig. 4B). All grids were thereafter treated identically.
Figure 4.
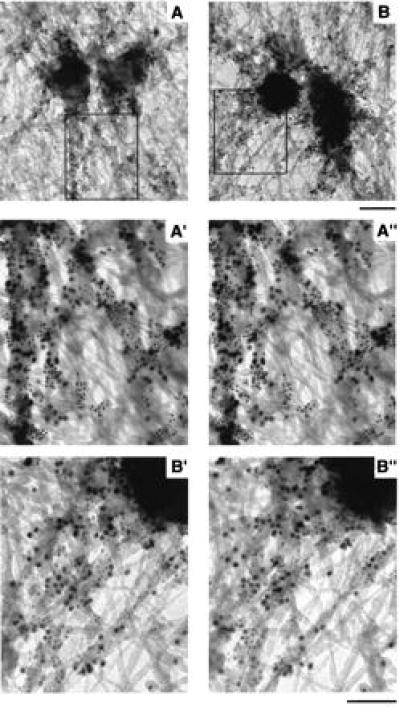
Immunogold localization of protein 4.1 and 5051 epitopes. In cell whole mounts of human fibroblasts, protein 4.1 epitopes (5-nm gold beads) and 5051 epitopes (10-nm gold beads) decorated fibrous structures in the centrosome region. A′ and A", and B′ and B" are stereo pairs of boxed regions in A and B. (A) Conditions that preserve microtubules. (B) Microtubule destabilizing conditions. (Upper bar = 200 nm; lower bar = 100 nm.)
Fig. 4 A and B shows protein 4.1 epitopes, labeled with 5-nm beads, on the outer surface of centriole cylinders and on fibrous projecting structures as in the previous single label experiments (Fig. 3). The distribution was independent of the presence (Fig. 4A) or absence (Fig. 4B) of microtubules. The 5051 epitopes, labeled by the 10-nm gold beads, were distributed similarly. Often both 4.1 and 5051 epitopes appeared on what seems to be a continuous filament. The relation between the epitopes is even clearer in the stereo pair images (Fig. 4 A′ and A", and B′ and B"). The 4.1 epitopes (5-nm beads) were distributed similarly to 5051 epitopes (10-nm beads), but there are many small domains apparently labeled by only one antibody.
Because 4.1 epitope localization is not markedly altered by the presence or absence of microtubules, it appears that most protein 4.1 epitopes do not colocalize with microtubules. We tested this prediction in double-labeling experiments using anti-4.1 and anti-β-tubulin antibodies under conditions for microtubule preservation. In Fig. 5, the dramatic pattern of fibers decorated with 10-nm beads (β-tubulin epitopes) radiating from the centrosomal region confirms that microtubules remained intact. By contrast, under microtubule destabilizing conditions, a fiber network was not labeled by anti-β-tubulin (data not shown). In microtubule preservation experiments (Fig. 5), protein 4.1 epitopes (5-nm beads) did not appear to colocalize with microtubules (10-nm beads). Lack of colocalization was also apparent in stereo images (data not shown). Rather, in a number of instances, 4.1 epitopes (5-nm beads) clustered in small areas between microtubules (Fig. 5, arrows).
Figure 5.
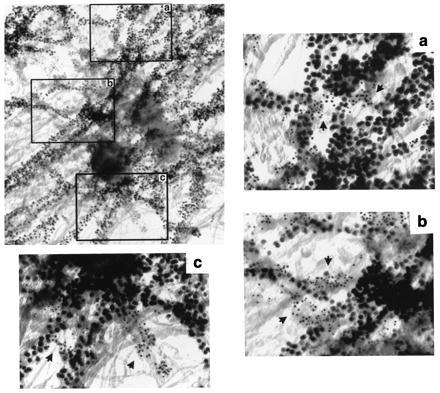
Cell whole mount electron micrograph of microtubules and protein 4.1 at a centrosome of a human fibroblast. Protein 4.1 epitopes (5-nm beads) and β-tubulin (10-nm beads) on microtubules were detected in fibroblasts prepared under conditions for microtubule stabilization. Three regions (a, b, and c), from the boxes on the image at low magnification, present examples of separate 4.1 epitope distribution (arrows) relative to microtubules. (Bar = 100 nm.)
DISCUSSION
Although long identified with organizing cell structure, the centrosome remains a puzzling organelle. It displays at least two impressive functions: the nucleation of microtubules and the apparent ability to replicate itself. Understanding these behaviors would seem to require detailed knowledge of structural components. Although the centrosomes examined here clearly consist of a centriole pair enmeshed in a surrounding matrix, little more is understood about the exact nature and extent of its structure. In this report we identify a new constituent of the centrosome, one or more members of the protein 4.1 family. We used antibodies to protein 4.1 and compared their staining patterns in optical and electron microscopy to that of the centrosome-specific 5051 autoimmune serum and anti-β-tubulin.
The centrosome is far too small to visualize in detail at the light microscope level. Electron microscopy, using the customary resin-embedded ultrathin sectioning of isolated human centrosomes, has revealed a symmetrical internal organization of centriole cylinders along with some PCM constituents such as centriole appendages, intercentriolar filament links, and satellites (29). Here we have used the technique of embedment-free electron microscopy to visualize the three-dimensional filament network surrounding the human centriole in situ. Detergent-extracted cell whole mounts are easily prepared and, with sufficiently thin cells, show the pericentriolar filaments in unprecedented detail. Immunogold labeling of the whole mounts affords a first step to identifying the micro domains that comprise the centriolar machinery.
We detected protein 4.1 at centrosomes of human and murine cells using affinity-purified antibodies directed against 80-kDa red cell protein 4.1 as well as against several 4.1 peptide domains. Similar results with multiple, independent 4.1 antibodies assures that authentic protein 4.1 or very highly related homologues of protein 4.1 were detected at centrosomes. The 4.1 epitopes were stably associated at centrosomes in cell samples extracted with detergent, salt, and DNase. Protein 4.1 epitopes were detected at centrosomes at all cell cycle stages but were rearranged during cell division (19). Notably, mitotic spindle poles were intensely stained, consistent with the fact that centrosomes are precursors to mitotic spindle poles.
Confocal light microscopy localized protein 4.1 to the PCM region of centrosomes. High resolution immunoelectron microscopy showed 4.1 epitopes were distributed on the outer surface of centrioles, on juxta-centriolar filaments, and on fibrous structures that presumably are constituents of the PCM. Gold beads bound to 4.1 antibody often displayed an irregular distribution along centriolar cylinders, decorating one end more heavily than the other. Other ultrastructural studies have demonstrated that centrioles display distinct organizations at their distal and proximal ends (20, 30, 31). We speculate that the 4.1 pattern could be related to a distal–proximal differentiation of centrioles.
Double-label immunoelectron microscopy showed that protein 4.1 and 5051 epitopes localize predominately, but not exclusively, to similar areas of the PCM. The labeling showed the two antibodies mostly decorate different small domains that are closely intermingled. Experiments probing for protein 4.1 in acentriolar centrosomes (28, 32–34) which retain PCM components, including the 5051 autoantigen pericentrin (28), may clarify whether 4.1 localization is independent of centrioles. Conversely, detailed localization of protein 4.1 in sperm may provide evidence for an association of 4.1 with sperm basal bodies which, in some species, are structurally related to centrioles and become incorporated into centrosomes after fertilization (23). Protein 4.1 has been reported in human sperm and, interestingly, defective expression of 4.1 resulted in severely amorphous sperm heads and infertility (35).
Deciphering the function of protein 4.1 in centrosomes may be aided by identifying specific protein 4.1 isoform(s) present in centrosomes. Alternative splicing of protein 4.1 mRNA may generate isoforms with domains specific for interactions with centrosomal proteins. Domains of 80-kDa red cell 4.1 which bind spectrin, actin, calmodulin, and myosin have been identified (reviewed in ref. 8). Centrosomal protein 4.1 isoforms may contain similar or unique domains with affinity for particular centrosomal residents. For example, because antibody against the spectrin-actin binding domain of protein 4.1 stains centrosomes, a potential protein 4.1 partner could be centractin (Arp-1), which shares ≈50% homology with actin (36, 37). Additional attractive candidates include centrosomal calmodulin and the related calcium binding protein centrin (38, 39). Similarly, because 4.1 binds the coiled coil protein myosin, possible interactions of 4.1 with centrosomal coiled coil proteins such as pericentrin (28), NuMA (40, 41), and ninein (42) are of interest. Such molecular interactions of protein 4.1 may ultimately be integral to mechanisms by which centrosomes organize the cytoskeletal network and direct mitosis.
Acknowledgments
We thank Dr. S. Lockett for confocal microscopy advice, Dr. J. A. Nickerson for helpful discussions, Dr. J. Campisi for HCA cells, Dr. S. Doxsey for 5051 serum, and Ms. A. Sheppard and Mr. D. Clark for preparation of this manuscript and its figures. This research was supported by Grant DK32094 from the National Institutes of Health.
ABBREVIATION
- PCM
pericentriolar material
References
- 1.Fairbanks G, Steck T L, Wallach D F. Biochemistry. 1971;10:2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- 2.Leto T L, Marchesi V T. J Biol Chem. 1984;259:4603–4608. [PubMed] [Google Scholar]
- 3.Conboy J G, Chan J, Mohandas N, Kan Y W. Proc Natl Acad Sci USA. 1988;85:9062–9065. doi: 10.1073/pnas.85.23.9062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conboy J G, Chan J, Chasis J A, Kan Y W, Mohandas N. J Biol Chem. 1991;266:8273–8280. [PubMed] [Google Scholar]
- 5.Tang T K, Leto T L, Correas I, Alonso M A, Marchesi V T, Benz E J., Jr Proc Natl Acad Sci USA. 1988;85:3713–3717. doi: 10.1073/pnas.85.11.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang T K, Quin Z, Marchesi V T, Benz E J. J Cell Biol. 1990;110:617–624. doi: 10.1083/jcb.110.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haltiwanger R S, Holt G D, Hart G W. J Biol Chem. 1990;265:2563–2568. [PubMed] [Google Scholar]
- 8.Conboy J G. Semin Hematol. 1993;30:58–73. [PubMed] [Google Scholar]
- 9.Anderson R A, Correas I, Mazzucco C, Castle J D, Marchesi V T. J Cell Biochem. 1988;37:269–284. doi: 10.1002/jcb.240370303. [DOI] [PubMed] [Google Scholar]
- 10.Granger B L, Lazarides E. Cell. 1984;37:595–607. doi: 10.1016/0092-8674(84)90390-8. [DOI] [PubMed] [Google Scholar]
- 11.Granger B L, Lazarides E. Nature (London) 1985;313:238–241. doi: 10.1038/313238a0. [DOI] [PubMed] [Google Scholar]
- 12.Cohen C M, Foley S F, Korsgen C. Nature (London) 1982;299:648–650. doi: 10.1038/299648a0. [DOI] [PubMed] [Google Scholar]
- 13.Lue R A, Marfatia S M, Branton D, Chishti A H. Proc Natl Acad Sci USA. 1994;91:9818–9822. doi: 10.1073/pnas.91.21.9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leto T L, Pratt B M, Madri J A. J Cell Physiol. 1986;127:423–431. doi: 10.1002/jcp.1041270311. [DOI] [PubMed] [Google Scholar]
- 15.Chasis J A, Coulombel L, Conboy J, McGee S, Andrews K, Kan Y, Mohandas N. J Clin Invest. 1993;91:329–338. doi: 10.1172/JCI116189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madri J A, Pratt B M, Yannariello-Brown J. In: Endothelial Cell-Extracellular Matrix Interactions. Simionescu N S, Simionescu M, editors. New York: Plenum; 1988. pp. 167–188. [Google Scholar]
- 17.Correas I. Biochem J. 1991;279:581–585. doi: 10.1042/bj2790581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Carcer G, Lallena M J, Correas I. Biochem J. 1995;312:871–877. doi: 10.1042/bj3120871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krauss S W, Larabell C A, Lockett S, Penman S, Mohandas N, Chasis J. J Cell Biol. 1997;137:275–289. doi: 10.1083/jcb.137.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borens M. In: Structure and Function of Isolated Centrosomes. Kalnins V I, editor. San Diego: Academic; 1992. pp. 2–37. [Google Scholar]
- 21.Moritz M, Braunfeld M B, Sedat J W, Alberts B, Agard D A. Nature (London) 1995;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- 22.Zheng Y, Wong M L, Alberts B, Mitchison T. Nature (London) 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
- 23.Balczon R. Int Rev Cytol. 1996;169:25–82. doi: 10.1016/s0074-7696(08)61984-1. [DOI] [PubMed] [Google Scholar]
- 24.Kalnins V I, editor. The Centrosome. San Diego: Academic; 1992. [Google Scholar]
- 25.Kellogg D R, Moritz M, Alberts B M. Annu Rev Biochem. 1994;63:639–674. doi: 10.1146/annurev.bi.63.070194.003231. [DOI] [PubMed] [Google Scholar]
- 26.Nickerson J A, Krockmalnic G, He D, Penman S. Proc Natl Acad Sci USA. 1990;87:2259–2263. doi: 10.1073/pnas.87.6.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pockwinse S, Krockmalnic G, Doxsey S J, Nickerson J, Lian J, Van Wijen A J, Stein J L, Stein G S, Penman S. Proc Natl Acad Sci USA. 1997;94:3022–3027. doi: 10.1073/pnas.94.7.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doxsey S J, Stein P, Evans L, Calarco P D, Kirschner M. Cell. 1994;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- 29.Paintrand M, Moudjou M, Delacroix H, Borens M. J Struct Biol. 1992;108:107–128. doi: 10.1016/1047-8477(92)90011-x. [DOI] [PubMed] [Google Scholar]
- 30.Komesli S, Job D, Paintrand M, Margolis R, Borens M. J Cell Biol. 1989;109:2869–2878. doi: 10.1083/jcb.109.6.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klotz C, Dabanvalle M C, Paintrand M, Weber H, Borens M, Karsenti E. J Cell Biol. 1990;110:405–415. doi: 10.1083/jcb.110.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szollozi D, Calarco P, Donahue R P. J Cell Biol. 1972;11:521–541. doi: 10.1242/jcs.11.2.521. [DOI] [PubMed] [Google Scholar]
- 33.Schatten G. Dev Biol. 1994;165:299–335. doi: 10.1006/dbio.1994.1256. [DOI] [PubMed] [Google Scholar]
- 34.Calarco-Gillam P D, Siebert M C, Hubble R, Mitchison T, Kirschner M. Cell. 1983;35:621–629. doi: 10.1016/0092-8674(83)90094-6. [DOI] [PubMed] [Google Scholar]
- 35.Rosseaux-Provost R, Lesur P, Collier F, Rigot J M, Dalla Venezia N, Saint Pol P, Delaunay J, Gauthier A, Rosseau J. Lancet. 1994;343:764–765. doi: 10.1016/s0140-6736(94)91840-6. [DOI] [PubMed] [Google Scholar]
- 36.Lees-Miller J P, Helfman D M, Schroer T A. Nature (London) 1992;359:244–246. doi: 10.1038/359244a0. [DOI] [PubMed] [Google Scholar]
- 37.Clark S W, Meyer D I. Nature (London) 1992;359:246–250. doi: 10.1038/359246a0. [DOI] [PubMed] [Google Scholar]
- 38.Errabolu R, Sanders M, Salisbury J. J Cell Sci. 1994;107:9–16. doi: 10.1242/jcs.107.1.9. [DOI] [PubMed] [Google Scholar]
- 39.Salisbury J. Curr Opin Cell Biol. 1995;7:39–45. doi: 10.1016/0955-0674(95)80043-3. [DOI] [PubMed] [Google Scholar]
- 40.Compton D A, Szilak I, Cleveland D W. J Cell Biol. 1992;116:1395–1408. doi: 10.1083/jcb.116.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang T K, Tang C-j, Chao Y-J, Wu C-W. J Cell Sci. 1994;107:1389–1402. doi: 10.1242/jcs.107.6.1389. [DOI] [PubMed] [Google Scholar]
- 42.Bouckson-Castaing V, Moudjou M, Ferguson D J, Mucklow S, Belkaid Y, Milon G. J Cell Sci. 1996;109:179–190. doi: 10.1242/jcs.109.1.179. [DOI] [PubMed] [Google Scholar]


