Abstract
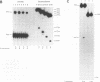
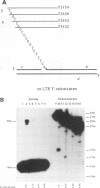
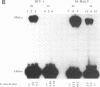
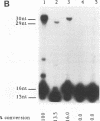
The disintegration activity of Moloney murine leukemia virus (M-MuLV) integrase (IN) was investigated through structural and sequence modifications of a Y substrate that resembles an integration intermediate. The Y substrates, constructed from individual oligonucleotides, contain a single viral long terminal repeat (LTR) joined to a nicked target DNA. Truncation of the double-stranded LTR sequences distal to the conserved 5'-CA-3' dinucleotide progressively diminished disintegration activity. M-MuLV IN was also able to catalyze disintegration of a heterologous double-stranded LTR sequence. Significantly, the activity of M-MuLV IN on single-stranded LTR Y substrates was more dependent on the sequence and length of the LTR strand than that reported for human immunodeficiency virus type 1 (HIV-1) IN. Modifications introduced at the Y-substrate junction demonstrated that the 3'-hydroxyl group at the terminus of the target strand was necessary for efficient joining of the target DNA strands. The presence of a 2'-hydroxyl group at the 3' end of the target strand, as well as a single-nucleotide gap at the LTR-target junction, reduced disintegration activity. The absence of hydroxyl groups on the terminal nucleotide abolished joining of the target strands. The results presented here suggest that M-MuLV IN disintegration activity is dependent on substantially different LTR sequence requirements than those reported for HIV-1 IN and may be mediated primarily through a structural recognition event.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown P. O., Bowerman B., Varmus H. E., Bishop J. M. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F. D., Craigie R. Sequence requirements for integration of Moloney murine leukemia virus DNA in vitro. J Virol. 1990 Nov;64(11):5645–5648. doi: 10.1128/jvi.64.11.5645-5648.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F. D., Engelman A., Palmer I., Wingfield P., Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F. D., Fujiwara T., Craigie R. Retroviral DNA integration directed by HIV integration protein in vitro. Science. 1990 Sep 28;249(4976):1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- Chow S. A., Vincent K. A., Ellison V., Brown P. O. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science. 1992 Feb 7;255(5045):723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- Colicelli J., Goff S. P. Mutants and pseudorevertants of Moloney murine leukemia virus with alterations at the integration site. Cell. 1985 Sep;42(2):573–580. doi: 10.1016/0092-8674(85)90114-x. [DOI] [PubMed] [Google Scholar]
- Colicelli J., Goff S. P. Sequence and spacing requirements of a retrovirus integration site. J Mol Biol. 1988 Jan 5;199(1):47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- Craigie R., Fujiwara T., Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990 Aug 24;62(4):829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- Engelman A., Mizuuchi K., Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991 Dec 20;67(6):1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Craigie R. Integration of mini-retroviral DNA: a cell-free reaction for biochemical analysis of retroviral integration. Proc Natl Acad Sci U S A. 1989 May;86(9):3065–3069. doi: 10.1073/pnas.86.9.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988 Aug 12;54(4):497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- Hong T., Murphy E., Groarke J., Drlica K. Human immunodeficiency virus type 1 DNA integration: fine structure target analysis using synthetic oligonucleotides. J Virol. 1993 Feb;67(2):1127–1131. doi: 10.1128/jvi.67.2.1127-1131.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto L. K., Halperin M., Champoux J. J. Moloney murine leukemia virus IN protein from disrupted virions binds and specifically cleaves its target sequence in vitro. Virology. 1991 Feb;180(2):527–534. doi: 10.1016/0042-6822(91)90066-k. [DOI] [PubMed] [Google Scholar]
- Johnson M. S., McClure M. A., Feng D. F., Gray J., Doolittle R. F. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson C. B., Donzella G. A., Roth M. J. Characterization of the forward and reverse integration reactions of the Moloney murine leukemia virus integrase protein purified from Escherichia coli. J Biol Chem. 1993 Jan 15;268(2):1462–1469. [PubMed] [Google Scholar]
- Jonsson C. B., Roth M. J. Role of the His-Cys finger of Moloney murine leukemia virus integrase protein in integration and disintegration. J Virol. 1993 Sep;67(9):5562–5571. doi: 10.1128/jvi.67.9.5562-5571.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz R. A., Merkel G., Kulkosky J., Leis J., Skalka A. M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990 Oct 5;63(1):87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- Katzman M., Katz R. A., Skalka A. M., Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol. 1989 Dec;63(12):5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan E., Mack J. P., Katz R. A., Kulkosky J., Skalka A. M. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 1991 Feb 25;19(4):851–860. doi: 10.1093/nar/19.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura Y., Lee Y. M., Coffin J. M. Nonrandom integration of retroviral DNA in vitro: effect of CpG methylation. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5532–5536. doi: 10.1073/pnas.89.12.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Kulkosky J., Skalka A. M. HIV DNA integration: observations and interferences. J Acquir Immune Defic Syndr. 1990;3(9):839–851. [PubMed] [Google Scholar]
- LaFemina R. L., Callahan P. L., Cordingley M. G. Substrate specificity of recombinant human immunodeficiency virus integrase protein. J Virol. 1991 Oct;65(10):5624–5630. doi: 10.1128/jvi.65.10.5624-5630.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt A. D., Rose R. B., Varmus H. E. Both substrate and target oligonucleotide sequences affect in vitro integration mediated by human immunodeficiency virus type 1 integrase protein produced in Saccharomyces cerevisiae. J Virol. 1992 Apr;66(4):2359–2368. doi: 10.1128/jvi.66.4.2359-2368.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara P. T., Bolshoy A., Trifonov E. N., Harrington R. E. Sequence-dependent kinks induced in curved DNA. J Biomol Struct Dyn. 1990 Dec;8(3):529–538. doi: 10.1080/07391102.1990.10507827. [DOI] [PubMed] [Google Scholar]
- Murphy J. E., Goff S. P. A mutation at one end of Moloney murine leukemia virus DNA blocks cleavage of both ends by the viral integrase in vivo. J Virol. 1992 Aug;66(8):5092–5095. doi: 10.1128/jvi.66.8.5092-5095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek E. Local supercoil-stabilized DNA structures. Crit Rev Biochem Mol Biol. 1991;26(2):151–226. doi: 10.3109/10409239109081126. [DOI] [PubMed] [Google Scholar]
- Pryciak P. M., Varmus H. E. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell. 1992 May 29;69(5):769–780. doi: 10.1016/0092-8674(92)90289-o. [DOI] [PubMed] [Google Scholar]
- Pullen K. A., Champoux J. J. Plus-strand origin for human immunodeficiency virus type 1: implications for integration. J Virol. 1990 Dec;64(12):6274–6277. doi: 10.1128/jvi.64.12.6274-6277.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M. J., Schwartzberg P. L., Goff S. P. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell. 1989 Jul 14;58(1):47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- Sherman P. A., Dickson M. L., Fyfe J. A. Human immunodeficiency virus type 1 integration protein: DNA sequence requirements for cleaving and joining reactions. J Virol. 1992 Jun;66(6):3593–3601. doi: 10.1128/jvi.66.6.3593-3601.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman P. A., Fyfe J. A. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci U S A. 1990 Jul;87(13):5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. A. DNA conformation and protein binding. Annu Rev Biochem. 1989;58:427–452. doi: 10.1146/annurev.bi.58.070189.002235. [DOI] [PubMed] [Google Scholar]
- Vincent K. A., Ellison V., Chow S. A., Brown P. O. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J Virol. 1993 Jan;67(1):425–437. doi: 10.1128/jvi.67.1.425-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink C., Yeheskiely E., van der Marel G. A., van Boom J. H., Plasterk R. H. Site-specific hydrolysis and alcoholysis of human immunodeficiency virus DNA termini mediated by the viral integrase protein. Nucleic Acids Res. 1991 Dec 25;19(24):6691–6698. doi: 10.1093/nar/19.24.6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink C., van Gent D. C., Plasterk R. H. Integration of human immunodeficiency virus types 1 and 2 DNA in vitro by cytoplasmic extracts of Moloney murine leukemia virus-infected mouse NIH 3T3 cells. J Virol. 1990 Oct;64(10):5219–5222. doi: 10.1128/jvi.64.10.5219-5222.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent D. C., Vink C., Groeneger A. A., Plasterk R. H. Complementation between HIV integrase proteins mutated in different domains. EMBO J. 1993 Aug;12(8):3261–3267. doi: 10.1002/j.1460-2075.1993.tb05995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]