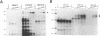Abstract
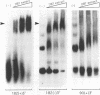
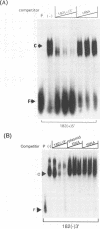
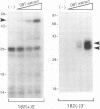
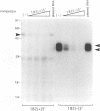
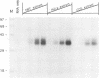
The termini of viral genomic RNA and its complementary strand are important in the initiation of viral RNA replication, which probably involves both viral and cellular proteins. To detect the possible cellular proteins involved in the replication of mouse hepatitis virus RNA, we performed RNA-protein binding studies with RNAs representing both the 5' and 3' ends of the viral genomic RNA and the 3' end of the negative-strand complementary RNA. Gel-retardation assays showed that both the 5'-end-positive- and 3'-end-negative-strand RNA formed an RNA-protein complex with cellular proteins from the uninfected cells. UV cross-linking experiments further identified a 55-kDa protein bound to the 5' end of the positive-strand viral genomic RNA and two proteins 35 and 38 kDa in size bound to the 3' end of the negative-strand cRNA. The results of the competition assay confirmed the specificity of this RNA-protein binding. No proteins were found to bind to the 3' end of the viral genomic RNA under the same conditions. The binding site of the 55-kDa protein was mapped within the 56-nucleotide region from nucleotides 56 to 112 from the 5' end of the positive-strand RNA, and the 35- and 38-kDa proteins bound to the complementary region on the negative-strand RNA. The 38-kDa protein was detected only in DBT cells but was not detected in HeLa or COS cells, while the 35-kDa protein was found in all three cell types. The juxtaposition of the different cellular proteins on the complementary sites near the ends of the positive- and negative-strand RNAs suggests that these proteins may interact with each other and play a role in mouse hepatitis virus RNA replication.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andino R., Rieckhof G. E., Baltimore D. A functional ribonucleoprotein complex forms around the 5' end of poliovirus RNA. Cell. 1990 Oct 19;63(2):369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- Baric R. S., Stohlman S. A., Lai M. M. Characterization of replicative intermediate RNA of mouse hepatitis virus: presence of leader RNA sequences on nascent chains. J Virol. 1983 Dec;48(3):633–640. doi: 10.1128/jvi.48.3.633-640.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claude A., Arenas J., Hurwitz J. The isolation and characterization of an RNA helicase from nuclear extracts of HeLa cells. J Biol Chem. 1991 Jun 5;266(16):10358–10367. [PubMed] [Google Scholar]
- Compton S. R., Rogers D. B., Holmes K. V., Fertsch D., Remenick J., McGowan J. J. In vitro replication of mouse hepatitis virus strain A59. J Virol. 1987 Jun;61(6):1814–1820. doi: 10.1128/jvi.61.6.1814-1820.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dildine S. L., Semler B. L. Conservation of RNA-protein interactions among picornaviruses. J Virol. 1992 Jul;66(7):4364–4376. doi: 10.1128/jvi.66.7.4364-4376.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhard J. R., Ehrenfeld E. Specific interactions of HeLa cell proteins with proposed translation domains of the poliovirus 5' noncoding region. J Virol. 1992 May;66(5):3101–3109. doi: 10.1128/jvi.66.5.3101-3109.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Hirano N., Fujiwara K., Hino S., Matumoto M. Replication and plaque formation of mouse hepatitis virus (MHV-2) in mouse cell line DBT culture. Arch Gesamte Virusforsch. 1974;44(3):298–302. doi: 10.1007/BF01240618. [DOI] [PubMed] [Google Scholar]
- Hirling H., Scheffner M., Restle T., Stahl H. RNA helicase activity associated with the human p68 protein. Nature. 1989 Jun 15;339(6225):562–564. doi: 10.1038/339562a0. [DOI] [PubMed] [Google Scholar]
- Joo M., Makino S. Mutagenic analysis of the coronavirus intergenic consensus sequence. J Virol. 1992 Nov;66(11):6330–6337. doi: 10.1128/jvi.66.11.6330-6337.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986 Sep 12;46(6):845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lai M. M. Coronavirus: organization, replication and expression of genome. Annu Rev Microbiol. 1990;44:303–333. doi: 10.1146/annurev.mi.44.100190.001511. [DOI] [PubMed] [Google Scholar]
- Lai M. M., Patton C. D., Baric R. S., Stohlman S. A. Presence of leader sequences in the mRNA of mouse hepatitis virus. J Virol. 1983 Jun;46(3):1027–1033. doi: 10.1128/jvi.46.3.1027-1033.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J., Shieh C. K., Gorbalenya A. E., Koonin E. V., La Monica N., Tuler J., Bagdzhadzhyan A., Lai M. M. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991 Feb;180(2):567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. J., Lai M. M. Deletion mapping of a mouse hepatitis virus defective interfering RNA reveals the requirement of an internal and discontiguous sequence for replication. J Virol. 1993 Oct;67(10):6110–6118. doi: 10.1128/jvi.67.10.6110-6118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luz N., Beck E. Interaction of a cellular 57-kilodalton protein with the internal translation initiation site of foot-and-mouth disease virus. J Virol. 1991 Dec;65(12):6486–6494. doi: 10.1128/jvi.65.12.6486-6494.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Joo M. Effect of intergenic consensus sequence flanking sequences on coronavirus transcription. J Virol. 1993 Jun;67(6):3304–3311. doi: 10.1128/jvi.67.6.3304-3311.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Lai M. M. High-frequency leader sequence switching during coronavirus defective interfering RNA replication. J Virol. 1989 Dec;63(12):5285–5292. doi: 10.1128/jvi.63.12.5285-5292.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Shieh C. K., Soe L. H., Baker S. C., Lai M. M. Primary structure and translation of a defective interfering RNA of murine coronavirus. Virology. 1988 Oct;166(2):550–560. doi: 10.1016/0042-6822(88)90526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesters H. G., Strauss J. H. Defined mutations in the 5' nontranslated sequence of Sindbis virus RNA. J Virol. 1990 Sep;64(9):4162–4168. doi: 10.1128/jvi.64.9.4162-4168.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachuk C. J., Bredenbeek P. J., Zoltick P. W., Spaan W. J., Weiss S. R. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis coronavirus, strain A59. Virology. 1989 Jul;171(1):141–148. doi: 10.1016/0042-6822(89)90520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardigon N., Strauss J. H. Cellular proteins bind to the 3' end of Sindbis virus minus-strand RNA. J Virol. 1992 Feb;66(2):1007–1015. doi: 10.1128/jvi.66.2.1007-1015.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Ries D., Bolger E., Chang L. J., Stoltzfus C. M. MHV nucleocapsid synthesis in the presence of cycloheximide and accumulation of negative strand MHV RNA. Virus Res. 1986 Dec;6(3):261–272. doi: 10.1016/0168-1702(86)90074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue G. P., Hall T. C. The requirement for a 5' stem-loop structure in brome mosaic virus replication supports a new model for viral positive-strand RNA initiation. J Virol. 1992 Feb;66(2):674–684. doi: 10.1128/jvi.66.2.674-684.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray B. K., Lawson T. G., Kramer J. C., Cladaras M. H., Grifo J. A., Abramson R. D., Merrick W. C., Thach R. E. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J Biol Chem. 1985 Jun 25;260(12):7651–7658. [PubMed] [Google Scholar]
- Rozen F., Edery I., Meerovitch K., Dever T. E., Merrick W. C., Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990 Mar;10(3):1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber E., Matthias P., Müller M. M., Schaffner W. Rapid detection of octamer binding proteins with 'mini-extracts', prepared from a small number of cells. Nucleic Acids Res. 1989 Aug 11;17(15):6419–6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh C. K., Soe L. H., Makino S., Chang M. F., Stohlman S. A., Lai M. M. The 5'-end sequence of the murine coronavirus genome: implications for multiple fusion sites in leader-primed transcription. Virology. 1987 Feb;156(2):321–330. doi: 10.1016/0042-6822(87)90412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan W., Delius H., Skinner M., Armstrong J., Rottier P., Smeekens S., van der Zeijst B. A., Siddell S. G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2(10):1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomori K., Banner L. R., Lai M. M. Heterogeneity of gene expression of the hemagglutinin-esterase (HE) protein of murine coronaviruses. Virology. 1991 Aug;183(2):647–657. doi: 10.1016/0042-6822(91)90994-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomori K., Lai M. M. Mouse hepatitis virus utilizes two carcinoembryonic antigens as alternative receptors. J Virol. 1992 Oct;66(10):6194–6199. doi: 10.1128/jvi.66.10.6194-6199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]