Abstract
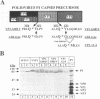
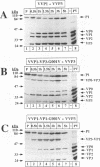
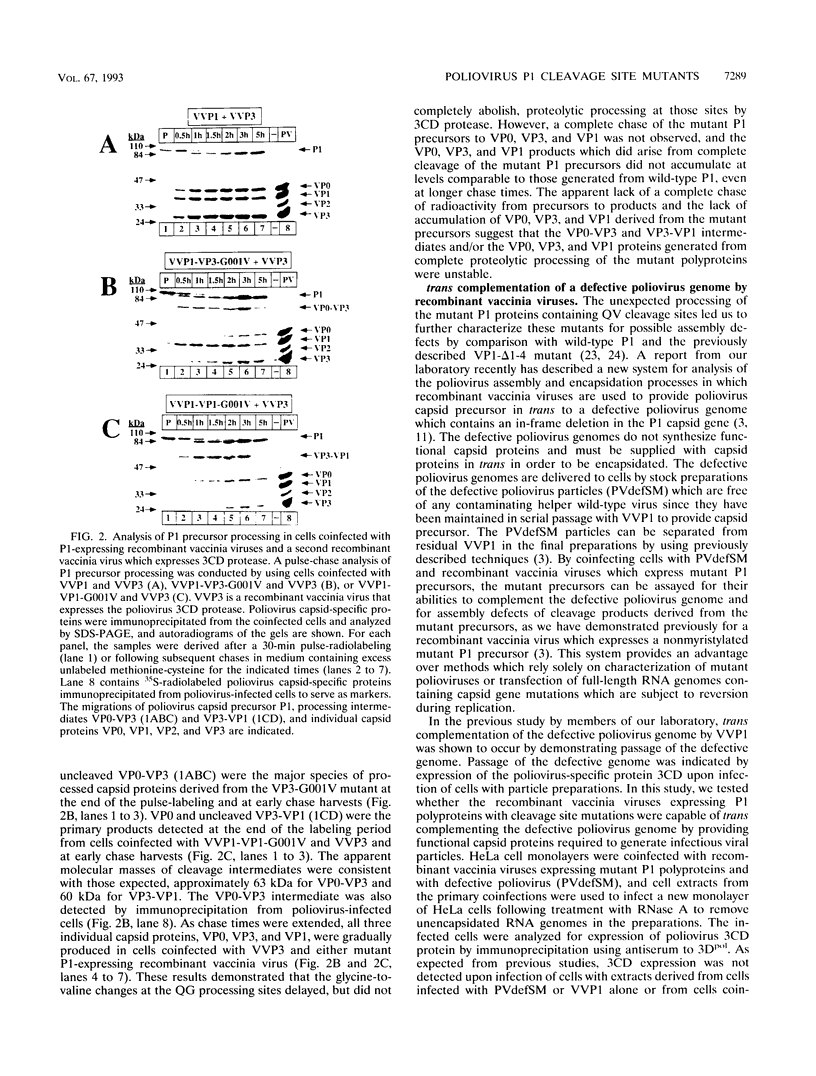
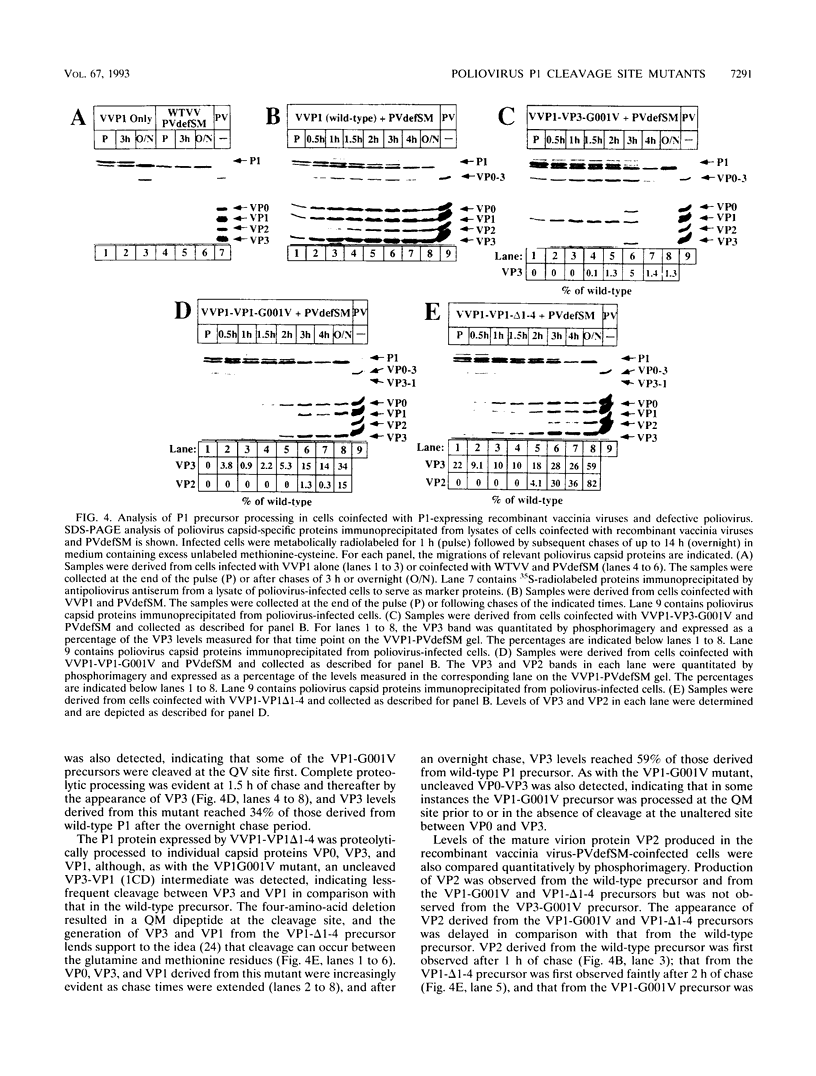
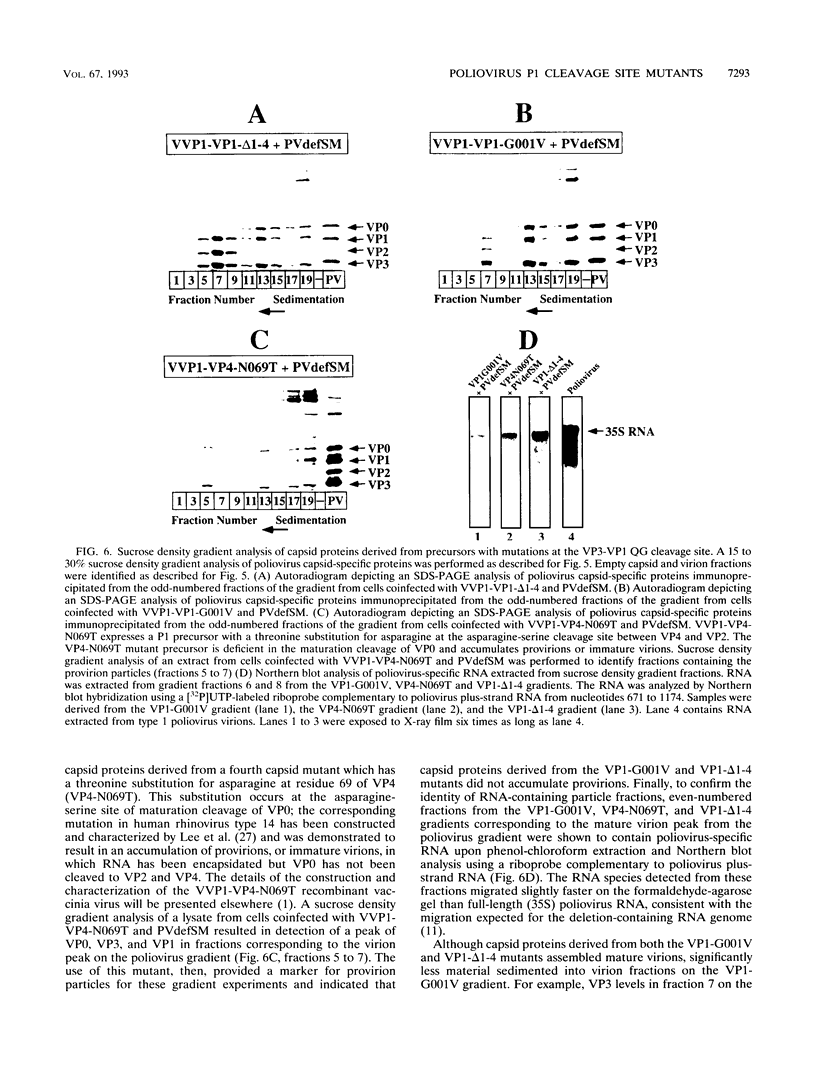
Assembly of poliovirus virions requires proteolytic cleavage of the P1 capsid precursor polyprotein between two separate glutamine-glycine (QG) amino acid pairs by the viral protease 3CD. In this study, we have investigated the effects on P1 polyprotein processing and subsequent assembly of processed capsid proteins caused by substitution of the glycine residue at the individual QG cleavage sites with valine (QG-->QV). P1 cDNAs encoding the valine substitutions were created by site-directed mutagenesis and were recombined into wild-type vaccinia virus to generate recombinant vaccinia viruses which expressed the mutant P1 precursors. The recombinant vaccinia virus-expressed mutant P1 polyproteins were analyzed for proteolytic processing defects in cells coinfected with a recombinant vaccinia virus (VVP3) that expresses the poliovirus 3CD protease and for processing and assembly defects by using a trans complementation system in which P1-expressing recombinant vaccinia viruses provide capsid precursor to a defective poliovirus genome that does not express functional capsid proteins (D. C. Ansardi, D. C. Porter, and C. D. Morrow, J. Virol. 67:3684-3690, 1993). The QV-substituted precursors were proteolytically processed at the altered sites both in cells coinfected with VVP3 and in cells coinfected with defective poliovirus, although the kinetics of cleavage at the altered sites were slower than those of cleavage at the wild-type QG site in the precursor. Completely processed capsid proteins VP0, VP3, and VP1 derived from the mutant precursor containing a valine at the amino terminus of VP3 (VP3-G001V) were unstable and failed to assemble stable subviral structures in cells coinfected with defective poliovirus. In contrast, capsid proteins derived from the P1 precursor with a valine substitution at the amino terminus of VP1 (VP1-G001V) assembled empty capsid particles but were deficient in assembling RNA-containing virions. The assembly characteristics of the VP1-G001V mutant were compared with those of a previously described VP3-VP1 cleavage site mutant (K. Kirkegaard and B. Nelsen, J. Virol. 64:185-194, 1990) which contained a deletion of the first four amino-terminal residues of VP1 (VP1-delta 1-4) and which was reconstructed for our studies into the recombinant vaccinia virus system. Complete proteolytic processing of the VP1-delta 1-4 precursor also occurred more slowly than complete cleavage of the wild-type precursor, and formation of virions was delayed; however, capsid proteins derived from the VP1-G001V mutant assembled RNA-containing virions less efficiently than those derived from the VP1-delta 1-4 precursor.(ABSTRACT TRUNCATED AT 400 WORDS)
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ansardi D. C., Porter D. C., Morrow C. D. Coinfection with recombinant vaccinia viruses expressing poliovirus P1 and P3 proteins results in polyprotein processing and formation of empty capsid structures. J Virol. 1991 Apr;65(4):2088–2092. doi: 10.1128/jvi.65.4.2088-2092.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansardi D. C., Porter D. C., Morrow C. D. Complementation of a poliovirus defective genome by a recombinant vaccinia virus which provides poliovirus P1 capsid precursor in trans. J Virol. 1993 Jun;67(6):3684–3690. doi: 10.1128/jvi.67.6.3684-3690.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansardi D. C., Porter D. C., Morrow C. D. Myristylation of poliovirus capsid precursor P1 is required for assembly of subviral particles. J Virol. 1992 Jul;66(7):4556–4563. doi: 10.1128/jvi.66.7.4556-4563.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold E., Luo M., Vriend G., Rossmann M. G., Palmenberg A. C., Parks G. D., Nicklin M. J., Wimmer E. Implications of the picornavirus capsid structure for polyprotein processing. Proc Natl Acad Sci U S A. 1987 Jan;84(1):21–25. doi: 10.1073/pnas.84.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair W. S., Semler B. L. Role for the P4 amino acid residue in substrate utilization by the poliovirus 3CD proteinase. J Virol. 1991 Nov;65(11):6111–6123. doi: 10.1128/jvi.65.11.6111-6123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S., Brechling K., Moss B. Vaccinia virus expression vector: coexpression of beta-galactosidase provides visual screening of recombinant virus plaques. Mol Cell Biol. 1985 Dec;5(12):3403–3409. doi: 10.1128/mcb.5.12.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain J. P. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979 Sep 15;98(1):132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- Choi W. S., Pal-Ghosh R., Morrow C. D. Expression of human immunodeficiency virus type 1 (HIV-1) gag, pol, and env proteins from chimeric HIV-1-poliovirus minireplicons. J Virol. 1991 Jun;65(6):2875–2883. doi: 10.1128/jvi.65.6.2875-2883.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricks C. E., Hogle J. M. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J Virol. 1990 May;64(5):1934–1945. doi: 10.1128/jvi.64.5.1934-1945.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagino-Yamagishi K., Nomoto A. In vitro construction of poliovirus defective interfering particles. J Virol. 1989 Dec;63(12):5386–5392. doi: 10.1128/jvi.63.12.5386-5392.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanecak R., Semler B. L., Anderson C. W., Wimmer E. Proteolytic processing of poliovirus polypeptides: antibodies to polypeptide P3-7c inhibit cleavage at glutamine-glycine pairs. Proc Natl Acad Sci U S A. 1982 Jul;79(13):3973–3977. doi: 10.1073/pnas.79.13.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen C. U., Wimmer E. Maturation of poliovirus capsid proteins. Virology. 1992 Apr;187(2):391–397. doi: 10.1016/0042-6822(92)90440-z. [DOI] [PubMed] [Google Scholar]
- Hellen C. U., Wimmer E. The role of proteolytic processing in the morphogenesis of virus particles. Experientia. 1992 Feb 15;48(2):201–215. doi: 10.1007/BF01923512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogle J. M., Chow M., Filman D. J. Three-dimensional structure of poliovirus at 2.9 A resolution. Science. 1985 Sep 27;229(4720):1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- Jablonski S. A., Luo M., Morrow C. D. Enzymatic activity of poliovirus RNA polymerase mutants with single amino acid changes in the conserved YGDD amino acid motif. J Virol. 1991 Sep;65(9):4565–4572. doi: 10.1128/jvi.65.9.4565-4572.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M. F., Asso J., Baltimore D. Further evidence on the formation of poliovirus proteins. J Mol Biol. 1970 May 14;49(3):657–669. doi: 10.1016/0022-2836(70)90289-5. [DOI] [PubMed] [Google Scholar]
- Jacobson M. F., Baltimore D. Morphogenesis of poliovirus. I. Association of the viral RNA with coat protein. J Mol Biol. 1968 Apr 28;33(2):369–378. doi: 10.1016/0022-2836(68)90195-2. [DOI] [PubMed] [Google Scholar]
- Jore J. P., Veldhuisen G., Pouwels P. H., Boeyé A., Vrijsen R., Rombaut B. Formation of subviral particles by in vitro translation of subgenomic poliovirus RNAs. J Gen Virol. 1991 Nov;72(Pt 11):2721–2726. doi: 10.1099/0022-1317-72-11-2721. [DOI] [PubMed] [Google Scholar]
- Jore J., De Geus B., Jackson R. J., Pouwels P. H., Enger-Valk B. E. Poliovirus protein 3CD is the active protease for processing of the precursor protein P1 in vitro. J Gen Virol. 1988 Jul;69(Pt 7):1627–1636. doi: 10.1099/0022-1317-69-7-1627. [DOI] [PubMed] [Google Scholar]
- Kean K. M., Teterina N., Girard M. Cleavage specificity of the poliovirus 3C protease is not restricted to Gln-Gly at the 3C/3D junction. J Gen Virol. 1990 Nov;71(Pt 11):2553–2563. doi: 10.1099/0022-1317-71-11-2553. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K. Mutations in VP1 of poliovirus specifically affect both encapsidation and release of viral RNA. J Virol. 1990 Jan;64(1):195–206. doi: 10.1128/jvi.64.1.195-206.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K., Nelsen B. Conditional poliovirus mutants made by random deletion mutagenesis of infectious cDNA. J Virol. 1990 Jan;64(1):185–194. doi: 10.1128/jvi.64.1.185-194.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura N., Semler B. L., Rothberg P. G., Larsen G. R., Adler C. J., Dorner A. J., Emini E. A., Hanecak R., Lee J. J., van der Werf S. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature. 1981 Jun 18;291(5816):547–553. doi: 10.1038/291547a0. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lee W. M., Monroe S. S., Rueckert R. R. Role of maturation cleavage in infectivity of picornaviruses: activation of an infectosome. J Virol. 1993 Apr;67(4):2110–2122. doi: 10.1128/jvi.67.4.2110-2122.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzayan C., Ingraham R., Wimmer E. Specificity of the polioviral proteinase 3C towards genetically engineered cleavage sites in the viral capsid. J Gen Virol. 1991 May;72(Pt 5):1159–1163. doi: 10.1099/0022-1317-72-5-1159. [DOI] [PubMed] [Google Scholar]
- Nicklin M. J., Harris K. S., Pallai P. V., Wimmer E. Poliovirus proteinase 3C: large-scale expression, purification, and specific cleavage activity on natural and synthetic substrates in vitro. J Virol. 1988 Dec;62(12):4586–4593. doi: 10.1128/jvi.62.12.4586-4593.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallai P. V., Burkhardt F., Skoog M., Schreiner K., Bax P., Cohen K. A., Hansen G., Palladino D. E., Harris K. S., Nicklin M. J. Cleavage of synthetic peptides by purified poliovirus 3C proteinase. J Biol Chem. 1989 Jun 15;264(17):9738–9741. [PubMed] [Google Scholar]
- Palmenberg A. C. In vitro synthesis and assembly of picornaviral capsid intermediate structures. J Virol. 1982 Dec;44(3):900–906. doi: 10.1128/jvi.44.3.900-906.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmenberg A. C. Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol. 1990;44:603–623. doi: 10.1146/annurev.mi.44.100190.003131. [DOI] [PubMed] [Google Scholar]
- Parks G. D., Palmenberg A. C. Site-specific mutations at a picornavirus VP3/VP1 cleavage site disrupt in vitro processing and assembly of capsid precursors. J Virol. 1987 Dec;61(12):3680–3687. doi: 10.1128/jvi.61.12.3680-3687.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnak J. R., Phillips B. A. Picornaviral structure and assembly. Microbiol Rev. 1981 Jun;45(2):287–315. doi: 10.1128/mr.45.2.287-315.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds C., Birnby D., Chow M. Folding and processing of the capsid protein precursor P1 is kinetically retarded in neutralization site 3B mutants of poliovirus. J Virol. 1992 Mar;66(3):1641–1648. doi: 10.1128/jvi.66.3.1641-1648.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda H., Nicklin M. J., Murray M. G., Anderson C. W., Dunn J. J., Studier F. W., Wimmer E. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell. 1986 Jun 6;45(5):761–770. doi: 10.1016/0092-8674(86)90790-7. [DOI] [PubMed] [Google Scholar]
- Wimmer E. Genome-linked proteins of viruses. Cell. 1982 Feb;28(2):199–201. doi: 10.1016/0092-8674(82)90335-x. [DOI] [PubMed] [Google Scholar]
- Ypma-Wong M. F., Dewalt P. G., Johnson V. H., Lamb J. G., Semler B. L. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988 Sep;166(1):265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]
- Ypma-Wong M. F., Filman D. J., Hogle J. M., Semler B. L. Structural domains of the poliovirus polyprotein are major determinants for proteolytic cleavage at Gln-Gly pairs. J Biol Chem. 1988 Nov 25;263(33):17846–17856. [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]