Abstract
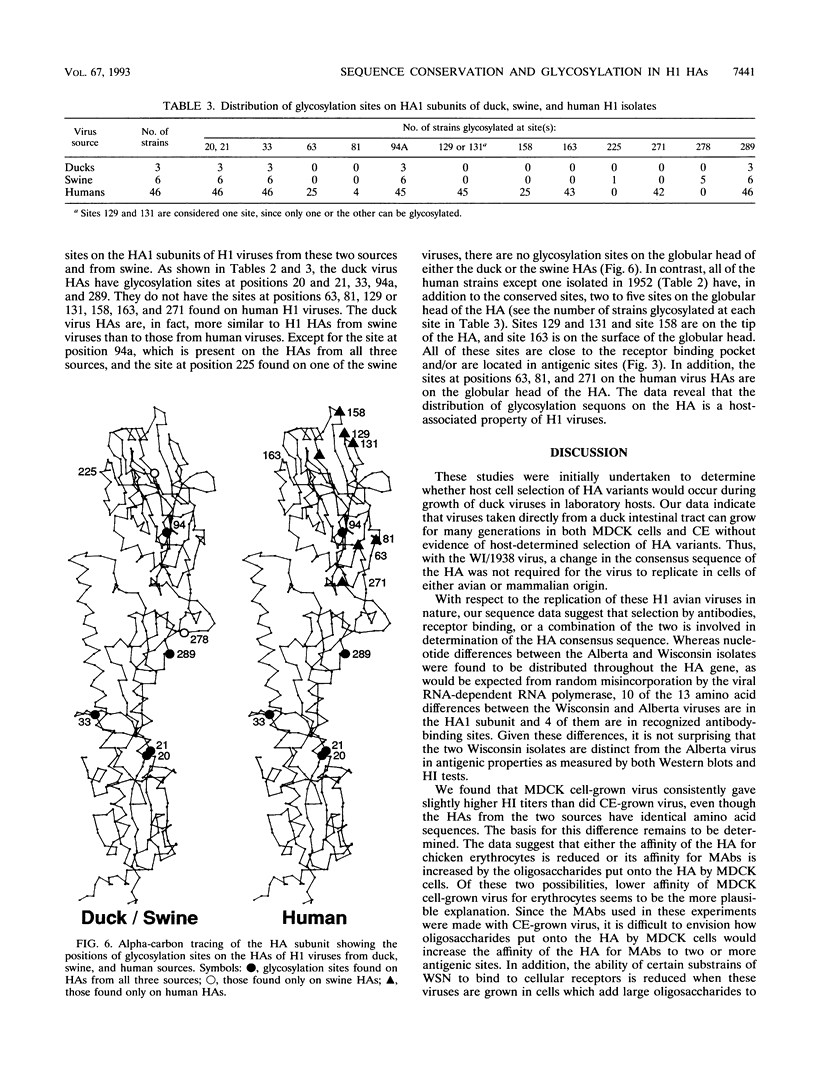
We determined the deduced amino acid sequences of two H1 duck influenza A virus hemagglutinins (HAs) and found that the consensus sequence of the HA, determined directly from virus recovered from the intestinal tract, remains unchanged through many generations of growth in MDCK cells and chicken embryos. These two duck viruses differ from each other by 5 amino acids and from A/Dk/Alberta/35/1976 (F. J. Austin, Y. Kawaoka, and R. G. Webster, J. Gen. Virol. 71:2471-2474, 1990) by 9 and 12 amino acids, most of which are in the HA1 subunit. They are antigenically similar to each other but different from the Alberta virus. We compared these H1 duck HAs with the HAs of human isolates to identify structural properties of this viral glycoprotein that are associated with host range. By comparison to the human H1 HAs, the duck virus HA sequences are highly conserved as judged by the small fraction of nucleotide differences between strains which result in amino acid substitutions. However, the most striking difference between these duck and human HAs is in the number and distribution of glycosylation sites. Whereas duck and swine viruses have four and five conserved glycosylation sites per HA1 subunit, none of which are on the tip of the HA, all human viruses have at least four additional sites, two or more of which are on the tip of the HA. These findings stress the role of glycosylation in the control of host range and suggest that oligosaccharides on the tip of the HA are important to the survival of H1 viruses in humans but not in ducks or swine.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Austin F. J., Kawaoka Y., Webster R. G. Molecular analysis of the haemagglutinin gene of an avian H1N1 influenza virus. J Gen Virol. 1990 Oct;71(Pt 10):2471–2474. doi: 10.1099/0022-1317-71-10-2471. [DOI] [PubMed] [Google Scholar]
- Austin F. J., Webster R. G. Antigenic mapping of an avian H1 influenza virus haemagglutinin and interrelationships of H1 viruses from humans, pigs and birds. J Gen Virol. 1986 Jun;67(Pt 6):983–992. doi: 10.1099/0022-1317-67-6-983. [DOI] [PubMed] [Google Scholar]
- Aytay S., Schulze I. T. Single amino acid substitutions in the hemagglutinin can alter the host range and receptor binding properties of H1 strains of influenza A virus. J Virol. 1991 Jun;65(6):3022–3028. doi: 10.1128/jvi.65.6.3022-3028.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beare A. S., Webster R. G. Replication of avian influenza viruses in humans. Arch Virol. 1991;119(1-2):37–42. doi: 10.1007/BF01314321. [DOI] [PubMed] [Google Scholar]
- Caton A. J., Brownlee G. G., Yewdell J. W., Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell. 1982 Dec;31(2 Pt 1):417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- Cox N. J., Black R. A., Kendal A. P. Pathways of evolution of influenza A (H1N1) viruses from 1977 to 1986 as determined by oligonucleotide mapping and sequencing studies. J Gen Virol. 1989 Feb;70(Pt 2):299–313. doi: 10.1099/0022-1317-70-2-299. [DOI] [PubMed] [Google Scholar]
- Crecelius D. M., Deom C. M., Schulze I. T. Biological properties of a hemagglutinin mutant of influenza virus selected by host cells. Virology. 1984 Nov;139(1):164–177. doi: 10.1016/0042-6822(84)90337-4. [DOI] [PubMed] [Google Scholar]
- Daniels P. S., Jeffries S., Yates P., Schild G. C., Rogers G. N., Paulson J. C., Wharton S. A., Douglas A. R., Skehel J. J., Wiley D. C. The receptor-binding and membrane-fusion properties of influenza virus variants selected using anti-haemagglutinin monoclonal antibodies. EMBO J. 1987 May;6(5):1459–1465. doi: 10.1002/j.1460-2075.1987.tb02387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deom C. M., Caton A. J., Schulze I. T. Host cell-mediated selection of a mutant influenza A virus that has lost a complex oligosaccharide from the tip of the hemagglutinin. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3771–3775. doi: 10.1073/pnas.83.11.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deom C. M., Schulze I. T. Oligosaccharide composition of an influenza virus hemagglutinin with host-determined binding properties. J Biol Chem. 1985 Nov 25;260(27):14771–14774. [PubMed] [Google Scholar]
- Hearing J., Gething M. J., Sambrook J. Addition of truncated oligosaccharides to influenza virus hemagglutinin results in its temperature-conditional cell-surface expression. J Cell Biol. 1989 Feb;108(2):355–365. doi: 10.1083/jcb.108.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw V. S., Bean W. J., Webster R. G., Sriram G. Genetic reassortment of influenza A viruses in the intestinal tract of ducks. Virology. 1980 Apr 30;102(2):412–419. doi: 10.1016/0042-6822(80)90108-7. [DOI] [PubMed] [Google Scholar]
- Hinshaw V. S., Webster R. G., Turner B. Novel influenza A viruses isolated from Canadian feral ducks: including strains antigenically related to swine influenza (Hsw1N1) viruses. J Gen Virol. 1978 Oct;41(1):115–127. doi: 10.1099/0022-1317-41-1-115. [DOI] [PubMed] [Google Scholar]
- Hiti A. L., Davis A. R., Nayak D. P. Complete sequence analysis shows that the hemagglutinins of the H0 and H2 subtypes of human influenza virus are closely related. Virology. 1981 May;111(1):113–124. doi: 10.1016/0042-6822(81)90658-9. [DOI] [PubMed] [Google Scholar]
- Katz J. M., Naeve C. W., Webster R. G. Host cell-mediated variation in H3N2 influenza viruses. Virology. 1987 Feb;156(2):386–395. doi: 10.1016/0042-6822(87)90418-1. [DOI] [PubMed] [Google Scholar]
- Katz J. M., Wang M., Webster R. G. Direct sequencing of the HA gene of influenza (H3N2) virus in original clinical samples reveals sequence identity with mammalian cell-grown virus. J Virol. 1990 Apr;64(4):1808–1811. doi: 10.1128/jvi.64.4.1808-1811.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka Y., Yamnikova S., Chambers T. M., Lvov D. K., Webster R. G. Molecular characterization of a new hemagglutinin, subtype H14, of influenza A virus. Virology. 1990 Dec;179(2):759–767. doi: 10.1016/0042-6822(90)90143-f. [DOI] [PubMed] [Google Scholar]
- Kida H., Kawaoka Y., Naeve C. W., Webster R. G. Antigenic and genetic conservation of H3 influenza virus in wild ducks. Virology. 1987 Jul;159(1):109–119. doi: 10.1016/0042-6822(87)90353-9. [DOI] [PubMed] [Google Scholar]
- Lu B. L., Webster R. G., Hinshaw V. S. Failure to detect hemagglutination-inhibiting antibodies with intact avian influenza virions. Infect Immun. 1982 Nov;38(2):530–535. doi: 10.1128/iai.38.2.530-535.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoh S. M., McGregor M. W., Hinshaw V. S. Hemagglutinin mutations related to antigenic variation in H1 swine influenza viruses. J Virol. 1992 Feb;66(2):1066–1073. doi: 10.1128/jvi.66.2.1066-1073.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANDEL B. Studies on the interactions of poliomyelitis virus, antibody, and host cells in tissue culture system. Virology. 1958 Oct;6(2):424–447. doi: 10.1016/0042-6822(58)90092-8. [DOI] [PubMed] [Google Scholar]
- Nohinek B., Gerhard W., Schulze I. T. Characterization of host cell binding variants of influenza virus by monoclonal antibodies. Virology. 1985 Jun;143(2):651–656. doi: 10.1016/0042-6822(85)90407-6. [DOI] [PubMed] [Google Scholar]
- Pensaert M., Ottis K., Vandeputte J., Kaplan M. M., Bachmann P. A. Evidence for the natural transmission of influenza A virus from wild ducts to swine and its potential importance for man. Bull World Health Organ. 1981;59(1):75–78. [PMC free article] [PubMed] [Google Scholar]
- Rajakumar A., Swierkosz E. M., Schulze I. T. Sequence of an influenza virus hemagglutinin determined directly from a clinical sample. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4154–4158. doi: 10.1073/pnas.87.11.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond F. L., Caton A. J., Cox N. J., Kendal A. P., Brownlee G. G. The antigenicity and evolution of influenza H1 haemagglutinin, from 1950-1957 and 1977-1983: two pathways from one gene. Virology. 1986 Jan 30;148(2):275–287. doi: 10.1016/0042-6822(86)90325-9. [DOI] [PubMed] [Google Scholar]
- Robertson J. S., Bootman J. S., Newman R., Oxford J. S., Daniels R. S., Webster R. G., Schild G. C. Structural changes in the haemagglutinin which accompany egg adaptation of an influenza A(H1N1) virus. Virology. 1987 Sep;160(1):31–37. doi: 10.1016/0042-6822(87)90040-7. [DOI] [PubMed] [Google Scholar]
- Rota P. A., Rocha E. P., Harmon M. W., Hinshaw V. S., Sheerar M. G., Kawaoka Y., Cox N. J., Smith T. F. Laboratory characterization of a swine influenza virus isolated from a fatal case of human influenza. J Clin Microbiol. 1989 Jun;27(6):1413–1416. doi: 10.1128/jcm.27.6.1413-1416.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheerar M. G., Easterday B. C., Hinshaw V. S. Antigenic conservation of H1N1 swine influenza viruses. J Gen Virol. 1989 Dec;70(Pt 12):3297–3303. doi: 10.1099/0022-1317-70-12-3297. [DOI] [PubMed] [Google Scholar]
- Skehel J. J., Stevens D. J., Daniels R. S., Douglas A. R., Knossow M., Wilson I. A., Wiley D. C. A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1779–1783. doi: 10.1073/pnas.81.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. F., Burgert E. O., Jr, Dowdle W. R., Noble G. R., Campbell R. J., Van Scoy R. E. Isolation of swine influenza virus from autopsy lung tissue of man. N Engl J Med. 1976 Mar 25;294(13):708–710. doi: 10.1056/NEJM197603252941308. [DOI] [PubMed] [Google Scholar]
- Underwood P. A., Skehel J. J., Wiley D. C. Receptor-binding characteristics of monoclonal antibody-selected antigenic variants of influenza virus. J Virol. 1987 Jan;61(1):206–208. doi: 10.1128/jvi.61.1.206-208.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R. G., Bean W. J., Gorman O. T., Chambers T. M., Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992 Mar;56(1):152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R. G., Yakhno M., Hinshaw V. S., Bean W. J., Murti K. G. Intestinal influenza: replication and characterization of influenza viruses in ducks. Virology. 1978 Feb;84(2):268–278. doi: 10.1016/0042-6822(78)90247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis W., Brown J. H., Cusack S., Paulson J. C., Skehel J. J., Wiley D. C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988 Jun 2;333(6172):426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- Winter G., Fields S., Brownlee G. G. Nucleotide sequence of the haemagglutinin gene of a human influenza virus H1 subtype. Nature. 1981 Jul 2;292(5818):72–75. doi: 10.1038/292072a0. [DOI] [PubMed] [Google Scholar]
- Xu X., Rocha E. P., Regenery H. L., Kendal A. P., Cox N. J. Genetic and antigenic analyses of influenza A (H1N1) viruses, 1986-1991. Virus Res. 1993 Apr;28(1):37–55. doi: 10.1016/0168-1702(93)90088-5. [DOI] [PubMed] [Google Scholar]
- Yewdell J. W., Caton A. J., Gerhard W. Selection of influenza A virus adsorptive mutants by growth in the presence of a mixture of monoclonal antihemagglutinin antibodies. J Virol. 1986 Feb;57(2):623–628. doi: 10.1128/jvi.57.2.623-628.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]





