Abstract
The c-Jun NH2-terminal kinase (JNK) group of mitogen-activated protein (MAP) kinases is activated by phosphorylation on Thr and Tyr. Here we report the molecular cloning of a new member of the mammalian MAP kinase kinase group (MKK7) that functions as an activator of JNK. In vitro protein kinase assays demonstrate that MKK7 phosphorylates and activates JNK, but not the p38 or extracellular signal-regulated kinase groups of MAP kinase. Expression of MKK7 in cultured cells causes activation of the JNK signal transduction pathway. MKK7 is therefore established to be a novel component of the JNK signal transduction pathway.
Three groups of mitogen-activated protein (MAP) kinases have been identified in mammals: the extracellular signal-regulated kinase (ERK); the p38 MAP kinase; and the c-Jun NH2-terminal kinase (JNK, also known as SAPK) (1). These MAP kinases are activated by dual phosphorylation within protein kinase subdomain VIII (1). This phosphorylation is mediated by a protein kinase cascade that consists of a MAP kinase, a MAP kinase kinase, and a MAP kinase kinase kinase. Individual MAP kinases are activated by different signaling modules that are regulated by different stimuli (2). For example, the ERK group is activated by the MAP kinase kinases MKK1 and MKK2; the p38 MAP kinase group is activated by MKK3, MKK4, and MKK6; and the JNK group is activated by MKK4 (1). These separate signaling modules allow the integrated response of MAP kinase pathways to different stimuli.
The JNK group of MAP kinases is activated by exposure of cells to environmental stress or by treatment of cells with pro-inflammatory cytokines (3–7). Targets of the JNK signal transduction pathway include the transcription factors ATF2 and c-Jun (1). These transcription factors are members of the basic leucine zipper (bZIP) group that bind as homo- and heterodimeric complexes to AP-1 and AP-1-like sites in the promoters of many genes (8). JNK binds to an NH2-terminal region of ATF2 and c-Jun and phosphorylates two sites within the activation domain of each transcription factor (4, 9–11). This phosphorylation leads to increased transcriptional activity (1). Together, these biochemical studies indicate that the JNK signal transduction pathway contributes to the regulation of AP-1 transcriptional activity in response to cytokines and environmental stress (1). Strong support for this hypothesis is provided by genetic evidence indicating that the JNK signaling pathway is required for the normal regulation of AP-1 transcriptional activity (12).
JNK is activated by dual phosphorylation on Thr-183 and Tyr-185 (4). MKK4 (also known as SEK1) is the only MAP kinase kinase that has been identified as a component of the JNK signal transduction pathway (2, 13, 14). Biochemical studies demonstrate that MKK4 phosphorylates and activates JNK (2, 13, 14). However, the function of MKK4 may not be restricted to the JNK signal transduction pathway because MKK4 also phosphorylates and activates p38 MAP kinase (2, 13). This specificity of MKK4 to activate both JNK and p38 MAP kinase provides a mechanism that may account for the coordinate activation of these MAP kinases in cells treated with cytokines or environmental stress (15). However, this coordinate activation is not always observed. For example, JNK activation in the liver correlates with decreased p38 MAP kinase activity (16). These data suggest that the properties of MKK4 are insufficient to account for the regulation of JNK in vivo.
The existence of a specific activator of JNK that is independent of MKK4 has been proposed previously (2). Indeed, biochemical studies support the conclusion that MKK4 is not the only activator of JNK in mammalian cells (17, 18). Furthermore, genetic evidence for this novel JNK activator was obtained from the results of experiments in which the MKK4 gene was disrupted by homologous recombination (12, 19). These studies demonstrate that although MKK4 (−/−) cells are defective in JNK regulation, the loss of MKK4 does not block JNK activation (12, 19). These studies provide definitive evidence that MKK4 represents only one mechanism of activation of the JNK protein kinase in vivo.
Here we describe the molecular cloning of a new member of the MAP kinase kinase group, MKK7. This protein kinase is widely expressed in mammalian tissues and, in contrast to MKK4, functions as a specific activator of JNK. We propose that MKK7 is a novel component of the JNK signal transduction pathway.
MATERIALS AND METHODS
Molecular Cloning of MKK7.
We designed the primers ATNGCNGTNAARCARATG and ATNCKYTCNGGNGCCATRTA based on the sequence of the Drosophila MAP kinase kinase hep (20). These primers were used in a reverse transcriptase–PCR (RT-PCR) with murine testis mRNA as the template. One sequence that encoded a hep-related protein kinase was identified. Full-length murine cDNA clones were isolated by screening a testis library (Stratagene). The cDNA clones were examined by sequencing with an Applied Biosystems model 373A machine. The MKK7 expression vector was constructed by subcloning a MKK7 cDNA (EcoRI and PvuII fragment) in the EcoRI and EcoRV sites of pCDNA3 (Invitrogen). The hep cDNA (20) was cloned by RT-PCR amplification of 0- to 4-hr Drosophila embryo mRNA (21).
Hybridization Analysis.
Northern blot analysis was done with poly(A)+ mRNA (2 μg) isolated from different tissues. The mRNA was fractionated by denaturing agarose gel electrophoresis and transferred to a nylon membrane (CLONTECH). The blot was probed with MKK4 and MKK7 cDNAs labeled by random priming with [α-32P]dATP (Amersham).
Recombinant Proteins.
The bacterial MKK7 expression vector was prepared by subcloning a MKK7 cDNA (EcoRI and PvuII fragment) in the EcoRI and SmaI sites of pGEX-5X1 (Pharmacia-LKB). The glutathione S-transferase (GST) fusion protein was purified by affinity chromatography (22). The recombinant proteins GST–ATF2 (10), GST–c-Jun (4), GST–c-Myc (23), GST–ERK2 (24), GST–p38α (25), and GST–JNK1 (4) have been described.
Tissue Culture.
CHO cells were maintained in DMEM supplemented with fetal calf serum (5%) (GIBCO/BRL). The cells were transfected with the lipofectamine reagent according to the manufacturer’s recommendations (GIBCO/BRL) (4).
Reporter Gene Expression.
Luciferase reporter gene expression was measured in cotransfection assays using 0.5 μg of the reporter plasmid pTRE-luciferase (26) and 0.25 μg of the β-galactosidase expression vector pCH110 (Pharmacia-LKB). Experiments using GAL4 fusion proteins were performed using 0.25 μg of pGAL4–ATF2 (residues 1–109), 0.5 μg of the reporter plasmid pG5E1bLuc, and 0.25 μg of pCH110 (10). The effect of protein kinases was examined by cotransfection with 0.3 μg of an empty expression vector or a protein kinase expression vector. The ERK2, p38α, JNK1, MKK1, MKK4, and MKK6 expression vectors have been described (2, 24, 25, 27). The cells were harvested 36 h post-transfection. The β-galactosidase and luciferase activity in the cell lysates was measured as described (10).
Immunoprecipitation.
The cells were solubilized with lysis buffer (20 mM Tris, pH 7.4/1% Triton X-100/10% glycerol/137 mM NaCl/2 mM EDTA/25 mM β-glycerophosphate/1 mM Na orthovanadate/2 mM pyrophosphate/1 mM phenylmethylsulfonyl fluoride/10 μg/ml leupeptin) and centrifuged at 100,000 × g for 15 min at 4°C. The epitope-tagged protein kinases were immunoprecipitated by incubation for 3 h at 4°C with an anti-HA monoclonal antibody bound to protein-G Sepharose (Pharmacia-LKB). The immunoprecipitates were washed three times with lysis buffer (10).
Protein Kinase Assays.
Protein kinase assays were performed in kinase buffer [25 mM 4-(2-hydroxyethyl)-1-piperazineethansulfonic acid, pH 7.4/25 mM β-glycerophosphate/25 mM MgCl2/2 mM dithiothreitol/0.1 mM orthovanadate]. The assays were initiated by the addition of 1 μg of substrate proteins and 50 μM [γ-32P]ATP (10 Ci/mmol; 1 Ci = 37 GBq) in a final volume of 25 μl. The reactions were terminated after 15 min at 25°C by addition of Laemmli sample buffer. The phosphorylation of the substrate proteins was examined after SDS/PAGE by autoradiography.
RESULTS
Molecular Cloning of MKK7.
We employed RT-PCR to identify a fragment of a novel mammalian MAP kinase kinase. The design of the primers was based on the sequence of the Drosophila MAP kinase kinase hep (20). The hep protein kinase is an activator of Drosophila JNK (21). A single product (461 bp) was detected following RT-PCR amplification of murine testis mRNA. Sequence analysis identified this PCR product as a fragment of a novel mammalian MAP kinase kinase. This cDNA fragment was used as a probe to screen a murine λ phage cDNA library to isolate full-length clones.
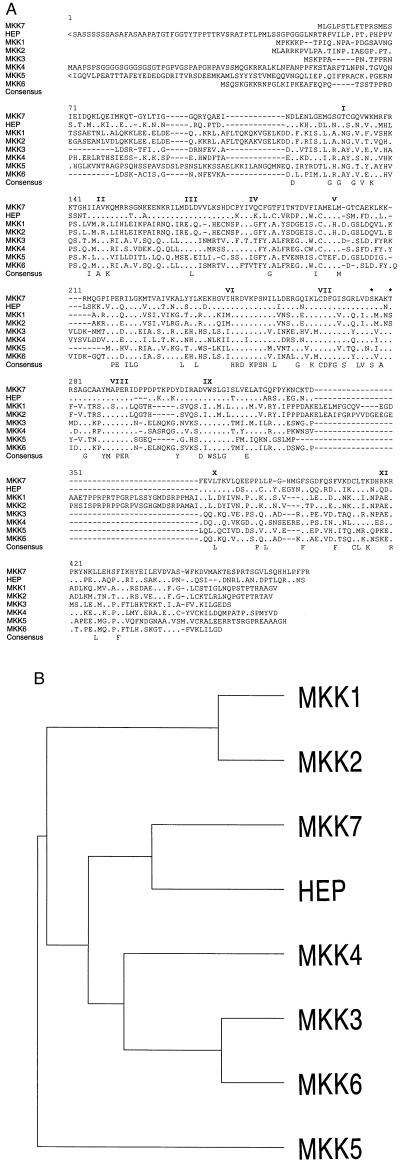
A group of seven clones was identified by sequence analysis to contain a single long open reading frame that encoded a putative protein kinase (Fig. 1). In-frame termination codons were detected in the 5′ and 3′ regions of these clones. This sequence includes protein kinase subdomains I–XI and is related to the MAP kinase kinase group. The novel protein kinase was designated MKK7. The sites of activating phosphorylation of MAP kinase kinases located in subdomain VIII are conserved in MKK7. Comparison of MKK7 with other members of the mammalian MAP kinase kinase group demonstrates that MKK7 is related to the JNK activator MKK4 (Fig. 1). MKK7 is most similar to the Drosophila JNK activator hep (Fig. 1)
Figure 1.
MKK7 is related to the Drosophila MAP kinase kinase hep. (A) The primary structure of MKK7 and HEP was deduced from the sequence of cDNA clones. These sequences were compared with the MAP kinase kinases MKK1, MKK2, MKK3, MKK4, MKK5, and MKK6 using the pile-up program (version 7.2; Wisconsin Genetics Computer Group, Madison). Gaps introduced into the sequences to optimize the alignment are illustrated with a dash (-). The sites of activating phosphorylation of MAP kinase kinases (2, 27–29) are indicated with asterisks (∗). The sequences of the MKK7, MKK7b, MKK7c, and hep cDNAs have been deposited in GenBank with accession numbers U93030, U93031, AF003199, and U93032, respectively. (B) The relationship between members of the MAP kinase kinase group (kinase subdomains I–XI) is presented as a dendrogram created by the unweighted pair-group method using arithmetic averages (pile-up program). The MAP kinase kinases MKK1, MKK2, MKK3, MKK4, MKK5, MKK6, and MKK7 and the Drosophila MAP kinase kinase hep are presented.
One additional cDNA clone isolated from the murine λ phage library differed from the other seven clones. This clone contained the same 3′-untranslated region and coding region of MKK7 but had a different 5′ region that lacked an in-frame termination codon. This clone may represent an alternatively spliced form of MKK7 (MKK7b). Similar forms of MKK1, MKK3, MKK4, MKK5, and MKK6 that are alternatively spliced in the 5′ region have been identified (2, 27, 30–33). The putative MKK7b cDNA clone we obtained does not have an initiation codon in the alternative 5′ region; this cDNA therefore encodes the same MKK7 protein kinase as the other clones we isolated. However, if the MKK7b cDNA clone is not full length, it is possible that additional 5′ sequence may include an in-frame initiation codon. If true, MKK7b is predicted to fuse the sequence M-[?]-SPAPAPSQRAALQLPLANDGGSRSPSSESSPQHPTPPTRPRH- to the initiating methionine of MKK7 (Fig. 1).
A human MKK7 cDNA was also identified. This cDNA contained an in-frame termination codon in the 5′-untranslated region and encoded a protein that fused the sequence MAASSLEQKLSRLEAKLKQENREARRRIDLNLDISPQRPRPTLQLPLANDGGSRSPSSESSPQHPTPPARPRH to the initiating methionine of MKK7. This NH2-terminal extension diverges from the sequence of mouse MKK7b. The human clone may therefore encode a different isoform, MKK7c. Together, these data indicate that the MKK7 gene is expressed as a group of protein kinases with different NH2 termini.
MKK7 Is Widely Expressed in Murine Tissues.
The expression of MKK7 was examined by Northern blot analysis of mRNA isolated from different tissues. MKK7 was found to be widely expressed in murine tissues. A single MKK7 transcript (≈4.0-kb) was detected in all of the tissues examined, except for testis where two MKK7 transcripts (4.0 and 1.6 kb) were detected (Fig. 2). MKK4 was also found to be widely expressed in murine tissues (Fig. 2). Although MKK4 and MKK7 are coexpressed, the relative abundance of each MAP kinase kinase is different in each of the tissues examined.
Figure 2.
MKK7 is widely expressed in mammalian tissues. The expression of MKK4 and MKK7 was examined by Northern blot analysis of poly(A)+ mRNA isolated from various murine tissues. Autoradiographs of the dried blots are presented.
MKK7 Is an Activator of JNK in Vitro.
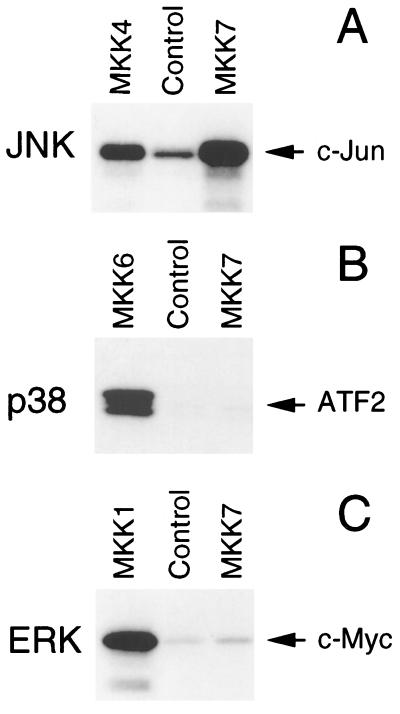
We performed in vitro protein kinase assays to examine the specificity of MKK7 (Fig. 3A). Recombinant MKK7 purified from bacteria was not observed to autophosphorylate. Incubation of the recombinant MKK7 with MAP kinases demonstrated that MKK7 phosphorylated JNK1, but not ERK2 or p38α. Control experiments demonstrated that ERK2 and p38α were activated by their cognate MAP kinase kinases, MKK1 and MKK6 (data not shown). Interestingly, MKK7 was phosphorylated by p38 and JNK1. The significance of this retrophosphorylation of the MAP kinase kinase by the MAP kinase is unclear, but similar retrophosphorylation has been detected in kinase assays using MKK4 (2) and the Drosophila JNK activator hep (21).
Figure 3.
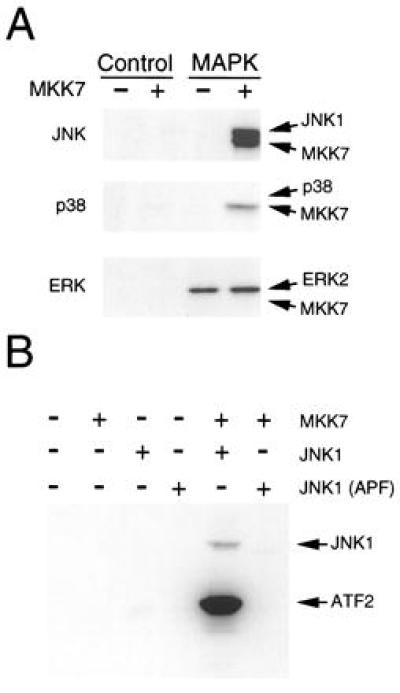
MKK7 is an activator of the JNK protein kinase in vitro. (A) Recombinant MAP kinases were incubated with GST (Control) or with GST–MKK7 in an in vitro protein kinase assay using the substrate ATP[γ-32P]. MKK7 phosphorylates JNK1, but not GST, ERK2, or p38α. MKK7 is phosphorylated by p38α and JNK1. (B) GST-MKK7 was incubated in a protein kinase assay with recombinant JNK1 or JNK1(APF). The mutated JNK1 protein (APF) was constructed by replacing the dual phosphorylation motif Thr-Pro-Tyr with Ala-Pro-Phe. The JNK activity was measured by including the JNK substrate ATF2 in each assay.
The test whether the phosphorylation of JNK1 by MKK7 caused increased protein kinase activity, we performed experiments using ATF2 as the JNK substrate (Fig. 3B). ATF2 was not phosphorylated by MKK7, but was weakly phosphorylated by JNK1. Incubation of MKK7 with JNK1 caused phosphorylation of JNK1 and a large increase in ATF2 phosphorylation. These data indicate that MKK7 phosphorylates and activates JNK1. To confirm this conclusion, we examined the effect of replacement of the JNK dual phosphorylation motif Thr-Pro-Tyr with Ala-Pro-Phe. The mutated JNK1 protein was not phosphorylated by MKK7. Furthermore, MKK7 did not increase ATF2 phosphorylation by the mutated JNK1 protein kinase. We conclude that MKK7 functions as an activator of JNK in vitro.
MKK7 Is an Activator of JNK in Vivo.
We performed cotransfection assays to examine the specificity of MKK7 in vivo. We found that the ERK2 and p38α MAP kinases were not activated by coexpressed MKK7 (Fig. 4A). Control experiments demonstrated that the ERK2 and p38α MAP kinases were activated by their respective cognate MAP kinase kinases, MKK1 and MKK6 (Fig. 4A). In contrast, MKK7 did activate JNK1 (Fig. 4A). Interestingly, the activation of JNK1 by coexpressed MKK7 was slightly greater than that caused by the previously described JNK activator MKK4. Control experiments using epitope-tagged activated MKK4 and MKK7 demonstrated that these MAP kinase kinases are expressed at similar levels, indicating that MKK7 may be a more potent activator of JNK than MKK4 (data not shown). Together, these data establish that MKK7 can function as an activator of JNK in cultured cells.
Figure 4.
MKK7 is an activator of the JNK protein kinase in vivo. (A) CHO cells were cotransfected with epitope-tagged JNK1 together with an empty expression vector (Control) or an expression vector encoding MKK4 or MKK7. The JNK1 was isolated by immunoprecipitation using an HA monoclonal antibody, and the protein kinase activity was measured in the immunecomplex with [γ-32P]ATP and c-Jun as substrates. The product of the phosphorylation reaction was visualized after SDS/PAGE by autoradiography. (B) The cells were cotransfected with epitope-tagged p38 MAP kinase together with an empty expression vector (Control) or an expression vector encoding MKK6 or MKK7. The activity of p38α MAP kinase was examined in an immunecomplex kinase assay using ATF2 as the substrate. (C) The cells were cotransfected with epitope-tagged ERK2 together with an empty expression vector (Control) or an expression vector encoding MKK1 or MKK7. The activity of ERK2 was examined in an immunecomplex kinase assay using c-Myc as the substrate.
JNK is activated by exposure of cells to environmental stress (4, 15). Exposure of transfected cells to UV-C radiation (80 J/m2) caused increased activity of both MKK7 and JNK (data not shown). MKK7 may therefore contribute to the regulation of JNK activity in vivo.
MKK7 Activates the JNK Signal Transduction Pathway.
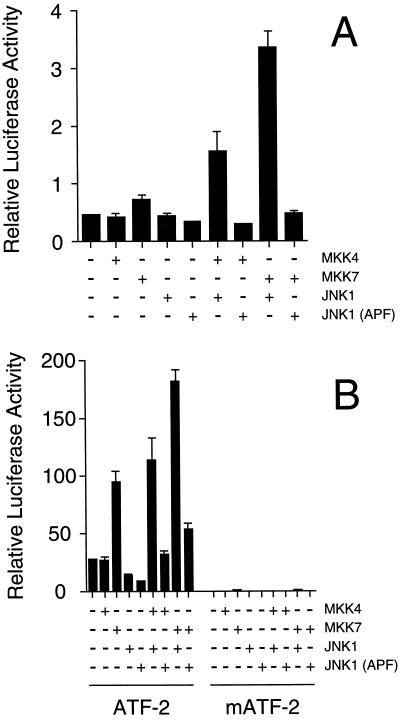
The JNK signaling pathway is known to regulate AP-1 transcriptional activity (1). We therefore postulated that the expression of MKK7 would cause increased AP-1 transcriptional activity. To test this hypothesis, we examined AP-1 transcriptional activity in a cotransfection assay employing a luciferase reporter gene that contains three AP-1 sites cloned upstream of a minimal promoter element (26). Expression of MKK4, MKK7, or JNK1 did not cause marked changes in AP-1 reporter gene expression (Fig. 5A). In contrast, coexpression of MKK7 with JNK1 caused increased AP-1-dependent reporter gene expression. Consistent with the observation that MKK4 causes a lower level of activation of JNK than MKK7 (Fig. 4), coexpression of MKK4 with JNK caused a smaller increase in AP-1 reporter gene expression (Fig. 5A). Together, these data demonstrate that MKK7 can function as an activator of the JNK signal transduction pathway.
Figure 5.
MKK7 is an activator of the JNK signal transduction pathway. (A) CHO cells were cotransfected with the AP-1 reporter plasmid pTRE-luciferase together with an empty expression vector, or expression vectors for MKK4, MKK7, JNK1, and JNK1 (APF). The mutated JNK1 protein (APF) was constructed by replacing the dual phosphorylation motif Thr-Pro-Tyr with Ala-Pro-Phe. Transfection efficiency was monitored by cotransfection with a β-galactosidase expression vector. The relative luciferase/β-galactosidase activity detected in the cell lysates is presented. The data shown are the mean ± SD (n = 3). (B) The transcriptional activity of a GAL4–ATF2 fusion protein was measured in a cotransfection assay in CHO cells using the reporter plasmid pG5E1bLuc. The effect of cotransfection with an empty expression vector or expression vectors for MKK4, MKK7, JNK1, and JNK1 (APF) was examined. Control experiments were performed using a mutated GAL4–ATF2 vector (mATF-2) in which the sites of ATF2 phosphorylation by JNK (Thr-69 and Thr-71) are replaced with Ala. Transfection efficiency was monitored by cotransfection with a β-galactosidase expression vector. The relative luciferase/β-galactosidase activity detected in the cell lysates is presented. The data shown are the mean ± SD (n = 3).
To further examine the effect of MKK7 on transcriptional activity, we investigated the effect of MKK7 on the transcription factor ATF2. Previous studies have demonstrated that ATF2 is a target of the JNK signal transduction pathway (9–11). JNK phosphorylates two sites (Thr-69 and Thr-71) in the NH2-terminal activation domain of ATF2 and increases transcriptional activity. We employed a GAL4 fusion protein strategy to monitor the transcriptional activity of the activation domain of ATF2 (10). Measurement of reporter gene expression demonstrated that the coexpression of MKK4 with JNK1 caused increased transcriptional activity (Fig. 5B). A similar level of reporter gene expression was caused by expression of MKK7 and a larger increase was detected when MKK7 was coexpressed with JNK1. The more potent effect of MKK7, compared with MKK4, on transcriptional activity is consistent with the relative effects of MKK7 and MKK4 on JNK activation (Fig. 4). To confirm that the increased reporter gene expression is mediated by ATF2 phosphorylation, we examined the effect of replacement of the sites of ATF2 phosphorylation (Thr-69 and Thr-71) with Ala. The mutated ATF2 protein was not regulated by MKK4, MKK7, or JNK1 (Fig. 5B). Together, these data demonstrate that MKK7 can regulate a physiological target of the JNK signaling pathway.
DISCUSSION
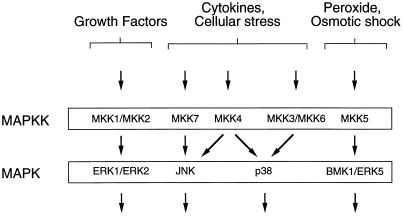
The mammalian MAP kinase kinase group includes at least seven members. The first group of MAP kinases that were identified are MKK1 and MKK2 (34). These protein kinases function as activators of the ERK group of MAP kinases (Fig. 6). Subsequent studies demonstrated that the p38 MAP kinases are activated by MKK3, MKK4, and MKK6; the BMK1/ERK5 MAP kinase is activated by MKK5; and the JNK group of MAP kinases is activated by MKK4 (1). Each of these MAP kinase kinases functions as an activator of a single group of MAP kinases with the exception of MKK4, which phosphorylates and activates both JNK and p38 MAP kinases (Fig. 6). A specific activator of JNK has therefore not been previously identified. Here we report the molecular cloning of a new member of the mammalian MAP kinase kinase group, MKK7. This MAP kinase kinase activates JNK, but not ERK2 or p38 MAP kinases in vitro (Fig. 3). Transfection assays demonstrate that MKK7 activates JNK and not the p38 or ERK MAP kinases in vivo (Fig. 4). The MKK7 protein kinase therefore appears to be the specific JNK activator that has been proposed in previous studies (2).
Figure 6.
MKK7 is an activator of the JNK signal transduction pathway. The ERK, BMK1/ERK5, p38, and JNK signal transduction pathways are illustrated schematically. MKK1 and MKK2 are activators of the ERK subgroup of MAP kinase. MKK3, MKK4, and MKK6 are activators of p38 MAP kinase. MKK5 is an activator of BMK1/ERK5. MKK7 is a specific activator of the JNK group of MAP kinases, whereas MKK4 activates both the p38 and JNK subgroups of MAP kinase.
MKK7 Is Related to the Drosophila MAP Kinase Kinase hep.
Comparison of the primary sequence of MKK7 with other members of the mammalian MAP kinase kinase group demonstrates that MKK7 is most similar to MKK4 (Fig. 1). This similarity in primary sequence reflects the common enzymatic property of MKK4 and MKK7 as activators of JNK (Fig. 6). However, MKK7 is most similar to the Drosophila MAP kinase kinase hep (Fig. 1). Biochemical analysis of hep demonstrates that it is a potent activator of Drosophila JNK in vitro (21). The function of hep as a physiological JNK activator is supported by genetic analysis. Loss-of-function alleles of the MAP kinase kinase (hep) and JNK (bsk) cause the same embryonic lethal phenotype (20, 21, 35). Detailed studies of Drosophila development demonstrate that the JNK protein kinase is required for morphogenetic cell movement during embryogenesis (20, 21, 35). Similarly, the JNK pathway is required for embryonic viability in mice (12). It is therefore possible that, like Drosophila, the JNK signaling pathway is required for embryonic morphogenesis in mammals. The specific targets of the JNK signaling pathway during embryonic development remain to be identified.
MKK7 and the JNK Signal Transduction Pathway.
The function of the JNK signal transduction pathway in vivo is poorly understood. Gene disruption experiments in mice demonstrate that the JNK pathway is required for the normal regulation of AP-1 transcriptional activity (12). The biological significance of AP-1 regulation by the JNK pathway is unclear, but this pathway has been implicated in the stress-induced apoptosis of neurons (36), the malignant transformation of pre-B cells (37), and the expression of E-selectin by endothelial cells (38). Further studies of the physiological role of the JNK signal transduction pathway in mammals will be greatly facilitated by the creation of animals with specific defects in JNK signaling. Targetted disruption of the MKK4 gene causes only a partial defect in JNK signaling (12, 19). It is likely that MKK7 may be able to complement the defect in MKK4 expression. The analysis of MKK4 knockout mice is therefore compromised by the widespread expression of MKK7 in murine tissues (Fig. 2). The identification of the MKK7 gene now allows the creation of murine cells that are defective in both of the known JNK activators, MKK4 and MKK7. These studies will assist our progress toward understanding the physiological function of the JNK signaling pathway.
Conclusions.
We report the molecular cloning of a new member of the MAP kinase kinase group in mammals, MKK7. This protein kinase is related the Drosophila MAP kinase kinase hep. Both hep and MKK7 function as activators of the JNK signal transduction pathway.
Acknowledgments
We thank K. Lei and I.-H. Wu for technical assistance and K. Gemme for administrative assistance. These studies were supported by Grant CA65861 from the National Cancer Institute. C.T. is an Institut National de la Santé et de la Recherche Médicale fellow and R.J.D. is an investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- MAP
mitogen-activated protein
- ERK
extracellular signal-regulated kinase
- JNK
c-Jun NH2-terminal kinase
- RT-PCR
reverse transcriptase–PCR
- GST
glutathione S-transferase
Footnotes
References
- 1.Whitmarsh A J, Davis R J. J Mol Med. 1996;74:589–607. doi: 10.1007/s001090050063. [DOI] [PubMed] [Google Scholar]
- 2.Dérijard B, Raingeaud J, Barrett T, Wu I-H, Han J, Ulevitch R J, Davis R J. Science. 1995;267:682–685. doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- 3.Gupta S, Barrett T, Whitmarsh A J, Cavanagh J, Sluss H K, Dérijard B, Davis R J. EMBO J. 1996;15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- 4.Dérijard B, Hibi M, Wu I-H, Barrett T, Su B, Deng T, Karin M, Davis R J. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 5.Kyriakis J M, Banerjee P, Nikolakaki E, Dai T, Rubie E A, Ahmad M F, Avruch J, Woodgett J R. Nature (London) 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 6.Sluss H K, Barrett T, Dérijard B, Davis R J. Mol Cell Biol. 1994;14:8376–8384. doi: 10.1128/mcb.14.12.8376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kallunki T, Su B, Tsigelny I, Sluss H K, Dérijard B, Moore G, Davis R J, Karin M. Genes Dev. 1994;8:2996–3007. doi: 10.1101/gad.8.24.2996. [DOI] [PubMed] [Google Scholar]
- 8.Curran T, Franza B J. Cell. 1988;55:395–397. doi: 10.1016/0092-8674(88)90024-4. [DOI] [PubMed] [Google Scholar]
- 9.van Dam H, Wilhelm D, Herr I, Steffen A, Herrlich P, Angel P. EMBO J. 1995;14:1798–1811. doi: 10.1002/j.1460-2075.1995.tb07168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta S, Campbell D, Dérijard B, Davis R J. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 11.Livingstone C, Patel G, Jones N. EMBO J. 1995;14:1785–1797. doi: 10.1002/j.1460-2075.1995.tb07167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang D, Tournier C, Wysk M, Lu H-T, Xu J, Davis R J, Flavell R A. Proc Natl Acad Sci USA. 1997;94:3004–3009. doi: 10.1073/pnas.94.7.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin A, Minden A, Martinetto H, Claret F-X, Lange-Carter C, Mercurio F, Johnson G L, Karin M. Science. 1995;268:286–290. doi: 10.1126/science.7716521. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez I, Hughes R T, Mayer B J, Yee K, Woodgett J R, Avruch J, Kyriakis J M, Zon L I. Nature (London) 1994;372:794–798. doi: 10.1038/372794a0. [DOI] [PubMed] [Google Scholar]
- 15.Davis R J. Trends Biochem Sci. 1994;19:470–473. doi: 10.1016/0968-0004(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 16.Mendelson K G, Contois L-R, Tevosian S G, Davis R J, Paulson K E. Proc Natl Acad Sci USA. 1996;93:12908–12913. doi: 10.1073/pnas.93.23.12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meier R, Rouse J, Cuenda A, Nebreda A R, Cohen P. Eur J Biochem. 1996;236:769–805. doi: 10.1111/j.1432-1033.1996.00796.x. [DOI] [PubMed] [Google Scholar]
- 18.Moriguchi T, Kawasaki H, Matsuda S, Gotoh Y, Nishida E. J Biol Chem. 1995;270:12969–12972. doi: 10.1074/jbc.270.22.12969. [DOI] [PubMed] [Google Scholar]
- 19.Nishina H, Fischer K D, Radvanyl L, Shahinian A, Hakem R, Ruble E A, Bernstein A, Mak T W, Woodgett J R, Penninger J M. Nature (London) 1997;385:350–353. doi: 10.1038/385350a0. [DOI] [PubMed] [Google Scholar]
- 20.Glise B, Bourbon H, Noselli S. Cell. 1995;83:451–461. doi: 10.1016/0092-8674(95)90123-x. [DOI] [PubMed] [Google Scholar]
- 21.Sluss H K, Han Z, Barrett T, Davis R J, Ip T. Genes Dev. 1996;10:2745–2758. doi: 10.1101/gad.10.21.2745. [DOI] [PubMed] [Google Scholar]
- 22.Smith S B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 23.Alvarez E, Northwood I C, Gonzalez F A, Latour D A, Seth A, Abate C, Curran T, Davis R J. J Biol Chem. 1991;266:15277–15285. [PubMed] [Google Scholar]
- 24.Seth A, Gonzalez F A, Gupta S, Raden D L, Davis R J. J Biol Chem. 1992;267:24796–24804. [PubMed] [Google Scholar]
- 25.Raingeaud J, Gupta S, Rogers J, Dickens M, Han J, Ulevitch R J, Davis R J. J Biol Chem. 1995;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 26.Rincon M, Flavell R A. EMBO J. 1994;13:4370–4381. doi: 10.1002/j.1460-2075.1994.tb06757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raingeaud J, Whitmarsh A J, Barrett T, Dérijard B, Davis R J. Mol Cell Biol. 1996;16:1247–1255. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alessi D R, Saito Y, Campbell D G, Cohen P, Sithanandam G, Rapp U, Ashworth A, Marshall C J, Cowley S. EMBO J. 1994;13:1610–1619. doi: 10.1002/j.1460-2075.1994.tb06424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng C F, Guan K L. EMBO J. 1994;13:1123–1131. doi: 10.1002/j.1460-2075.1994.tb06361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moriguchi T, Toyoshima F, Gotoh Y, Iwamatsu A, Irie K, Mori E, Kuroyanagi N, Hagiwara M, Matsumoto K, Nishida E. J Biol Chem. 1996;271:26981–26988. doi: 10.1074/jbc.271.43.26981. [DOI] [PubMed] [Google Scholar]
- 31.English J M, Vanderbilt C A, Xu S, Marcus S, Cobb M H. J Biol Chem. 1995;270:28897–28902. doi: 10.1074/jbc.270.48.28897. [DOI] [PubMed] [Google Scholar]
- 32.Seger R, Seger D, Lozeman F J, Ahn N G, Graves L M, Campbell J S, Ericsson L, Harrylock M, Jensen A M, Krebs E G. J Biol Chem. 1992;267:25628–25631. [PubMed] [Google Scholar]
- 33.Han J, Lee J-D, Jiang Y, Li Z, Feng L, Ulevitch R J. J Biol Chem. 1996;271:2886–2891. doi: 10.1074/jbc.271.6.2886. [DOI] [PubMed] [Google Scholar]
- 34.Ahn N G, Seger R, Krebs E G. Curr Opin Cell Biol. 1992;4:992–999. doi: 10.1016/0955-0674(92)90131-u. [DOI] [PubMed] [Google Scholar]
- 35.Riesgo-Escovar J R, Jenni M, Fritz A, Hafen E. Genes Dev. 1996;10:2759–2768. doi: 10.1101/gad.10.21.2759. [DOI] [PubMed] [Google Scholar]
- 36.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 37.Raitano A B, Halpern J R, Hambuch T M, Sawyers C L. Proc Natl Acad Sci USA. 1995;92:11746–11750. doi: 10.1073/pnas.92.25.11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Read M A, Whitely M Z, Gupta S, Pierce J W, Best J, Davis R J, Collins T. J Biol Chem. 1997;272:2753–2761. doi: 10.1074/jbc.272.5.2753. [DOI] [PubMed] [Google Scholar]