Abstract
The twn2 mutant of Arabidopsis exhibits a defect in early embryogenesis where, following one or two divisions of the zygote, the decendents of the apical cell arrest. The basal cells that normally give rise to the suspensor proliferate abnormally, giving rise to multiple embryos. A high proportion of the seeds fail to develop viable embryos, and those that do, contain a high proportion of partially or completely duplicated embryos. The adult plants are smaller and less vigorous than the wild type and have a severely stunted root. The twn2-1 mutation, which is the only known allele, was caused by a T-DNA insertion in the 5′ untranslated region of a putative valyl-tRNA synthetase gene, valRS. The insertion causes reduced transcription of the valRS gene in reproductive tissues and developing seeds but increased expression in leaves. Analysis of transcript initiation sites and the expression of promoter–reporter fusions in transgenic plants indicated that enhancer elements inside the first two introns interact with the border of the T-DNA to cause the altered pattern of expression of the valRS gene in the twn2 mutant. The phenotypic consequences of this unique mutation are interpreted in the context of a model, suggested by Vernon and Meinke [Vernon, D. M. & Meinke, D. W. (1994) Dev. Biol. 165, 566–573], in which the apical cell and its decendents normally suppress the embryogenic potential of the basal cell and its decendents during early embryo development.
In Arabidopsis and many other higher plants, the unfertilized egg cell is highly polarized. It contains a large central vacuole asymmetrically positioned proximal to the micropyle, whereas the nucleus and most of the cytoplasm are at the chalazal end (1). After fertilization, the zygote elongates along the future shoot–root axis and a redistribution of the endoplasmic reticulum, plastids, and mitochondria accentuates the polar organization evident in the egg cell (2, 3). Embryogenesis is initiated when the zygote undergoes an asymmetric transverse division to produce a relatively small, nonvacuolated apical cell, and a larger highly vacuolated basal cell (1, 4). The apical cell undergoes many rounds of organized cell division and gives rise to the embryo. The basal cell divides only a few times, always in the transverse orientation, forming the suspensor, the root cap, and a portion of the root meristem (5). The suspensor, which connects the maternal tissue and the growing embryo and is thought to be a source of growth factors in some species, eventually deteriorates and is not present in the mature embryo (6, 7).
Several mechanisms have been proposed to explain how the two daughter cells of the zygote follow different fates. One proposal is that cell identity might be determined by the relative position of the two cells in the embryo sac (8). For instance, it is conceivable the cell that is in contact with the maternal tissue always becomes the suspensor. Implicit in this hypothesis is the idea that the daughter cells communicate with each other or the maternal tissue to establish cell fate. The observation that suspensors do not develop during somatic embryogenesis seems consistent with this hypothesis (9, 10). An alternative proposal is that asymmetric division generates daughter cells with different sizes, and this difference may be sufficient to direct cell fates (11). A third possibility is that the daughter cells may inherit different cytoplasmic determinants of either zygotic or maternal origin that control cell fate (12).
Several Arabidopsis mutants with abnormal suspensor development have been described. The raspberry1 and raspberry2 mutants remain globular shaped, fail to form an axis and cotyledons, and have an enlarged multi-tiered suspensor region (13). Similarly, the abnormal suspensor mutants (sus1, sus2, and sus3) are characterized by a morphologically abnormal embryo that fails to form a viable seedling but has an enlarged suspensor (14). Because the raspberry and sus mutations cause the arrest of the embryo-proper, it could be argued that the abnormal suspensor development in these mutants results from the absence of a viable embryo-proper. In contrast, in the twn1 mutant, the embryo proper usually develops relatively normally, and the primary embryo, as well as the supernumerary embryos derived from the suspensor, are able to survive desiccation and produce viable seedlings. Thus, Vernon and Meinke (15) suggested that the TWN1 gene product is involved in maintaining suspensor cell identity, and mutations in the gene relieve the inhibitory effect normally imposed on the suspensor by the embryo.
Here, we describe a second Arabidopsis mutant that produces a high frequency of twin embryos and partial duplications of organs. This phenotype results from arrested development of the apical cells, followed by abnormal proliferation of the basal cells to produce secondary embryos. The properties of this mutant are consistent with a proposal by Vernon and Meinke (15) that the apical cell normally interacts with the basal cell to determine proper cell fates by suppressing the embryonic potential of the basal cell.
MATERIALS AND METHODS
Plant Materials and Mutant Screen.
Pools of T-DNA mutagenized seeds of the WS ecotype (16) were obtained from the Arabidopsis Biological Research Center (Columbus, OH) and screened for abnormal seed morphology by visual inspection of mature seeds under a dissecting microscope. Arabidopsis plants were grown in a glass house with ≈16-h light and 8-h dark photoperiod at an average temperature of 22°C. The mutant was backcrossed twice to the WS parent before being used for the experiments reported here.
Light Microscopy of Cleared Seeds.
Whole seeds at various stages of development were soaked in Histochoice tissue fixative (Amresco, Solon, OH) for 30 min and transferred to a drop of Hoyer’s solution (7.5 g gum arabic/100 g chloral hydrate/5 ml glycerol in 30 ml water) and cleared overnight. Cleared seeds were visualized using a Leica LEITZ DMRB microscope equipped with Nomarski optics.
Light Microscopy of Sections.
Plant material was fixed in 3% (vol/vol) glutaraldehyde in 50 mM sodium phosphate buffer (pH 7.4), at ambient temperature for 3 h with one change of fixative after 1.5 h. After rinsing in the same phosphate buffer four times (20 min each rinse), the material was dehydrated in a graded ethanol series to 100%, pre-infiltrated with 1:1 (vol/vol) mixture of ethanol and Spurr’s epoxy resin over a period of 48 h, infiltrated in pure Spurr’s resin for an additional 48 h, and embedded in fresh Spurr’s resin. Polymerization was carried out at 70°C overnight. Sections (2–3 μm) were cut with a Jung RM2025 microtome (Leica). Sections were stained with 0.05% analine blue for 1 h, rinsed with water, and visualized with a Leica LEITZ DMRB microscope.
Recombinant DNA Methods.
A genomic library of wild-type (Columbia ecotype) and λZAPII cDNA libraries representing siliques (WS ecotype) and flowers (Ler ecotype) were obtained from the Arabidopsis Biological Research Center at Ohio State University. A genomic library of the mutant was constructed from DNA digested with HindIII, blunted with Klenow, and ligated to EcoRI adapters (Promega). The adapters were removed on a sephacryl S-500 spin column (Stratagene), and the DNA was ligated to EcoRI-digested ZAPII arms (Stratagene). Plaque lifts were screened with the left border of the T-DNA. DNA sequences were obtained using an Applied Biosystems automated sequencer (ABI PRISM310).
The longest cDNA clone containing the valyl-tRNA synthetase (valRS) gene from the Columbia ecotype had a 3.6-kb insert and was designated pVRS19. Probes for hybridization experiments were labeled with 32P by random priming. The probe for Northern blot analyses was a 3.6-kb ApaI–SacI fragment from pVRS19. The Northern blots contained 10 μg total RNA per lane, hybridized in 5× SSPE (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA), 5× Denhardt’s solution, 50% formamide, and 1% SDS at 42°C. The 5′ rapid amplification of cDNA ends (RACE) procedure was done following the manufacturer’s instructions (BRL), except that a third nested primer was used (instead of only two). The sequences of the primers were 5′-CCGGGTACCCACAAAGCATT-3′, 5′-GTATCCTCAATTGCAGATGTAAGCG-3′, and 5′-ATATGCAAGGCTCCAGTCAC-3′.
Plant Transformation.
For complementation experiments, genomic fragments were cloned into vector pBIN-HYG (17), which contains a hygromycin resistance (hygr) gene. Homozygous twn2 plants were transformed using the in planta transformation method (18). Briefly, bolts were cut off with a razor blade and a drop of Agrobacterium suspension (10 ml overnight culture resuspended in 0.5 ml Murashige and Skoog medium) was put on the wound surface. Plants were then allowed to set seeds. Seeds were surface sterilized with 30% bleach and plated on agar media containing Murashige and Skoog salts (Sigma) and 25 mg/liter kanamycin and 30 mg/liter hygromycin. Heterozygous twn2 plants were transformed following the vacuum infiltration method (19).
For the construction of the promoter-reporter constructs pGUS1 and pGUS2, the promoter fragments were obtained by PCR using genomic clone pSac9.0 as template. The downstream primers in the PCRs for pGUS1 and pGUS2 were 5′-CGG GAT CCA TAG ACA AAA AAC CTG CTA GGG C-3′ and 5′-CGG GAT CCA ACT CCT TTT CTT TAG CCT ATG, respectively. Both primers had BamHI sites added to the 5′ end. The upstream primer for pGUS1 and pGUS2 was 5′-GTG TGG GTA ACC AAA GAC G-3′. The PCR products were first cloned into the pGEM-T vector (Promega), isolated as SalI/BamHI fragments, and ligated into the SalI/BamHI sites of the binary vector pBI101.2, which contains the uidA gene and the NOS terminator. Wild-type Arabidopsis (Columbia ecotype) was transformed as described (19).
RESULTS
Genetic Properties of the twn2 Mutant.
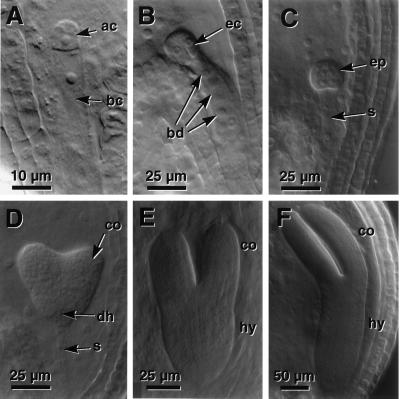
The twn2 mutant was isolated by visually screening ≈200 mature seeds from each of 120 pools representing 6,500 F3 families of T-DNA insertions for mutations that affected seed morphology. The homozygous twn2 line, recovered from pool CS2644, had deformed seeds (Fig. 1A), of which ≈80% did not germinate. Microscopic inspection of the nongerminated seeds indicated that most contained aborted embryos with abnormal proliferation of cells in place of suspensors. Among the germinated seeds, about 40% (or 37/92) produced seedlings that were partially or completely duplicated (Fig. 1 B–D). Because many seeds produced two complete seedlings, we refer to this somewhat variable phenotype as twin. The viable seeds also had a high incidence of seedlings with one or three cotyledons. All seedlings had stunted roots (Fig. 1 B–D) and generally developed more slowly than wild type. twn2 adult plants from a twice backcrossed line were less vigorous and smaller than wild type, had smaller and rounder leaves, and were less fertile (Fig. 1 E).
Figure 1.

Wild-type and twn2 seeds, seedlings, and adult plants. (A) Wild-type and twn2 seeds. (B–D) twn2 seedlings. (E) Wild-type and twn2 adult plants.
In a cross of twn2 and wild type, all 110 F1 seeds were kanamycin resistant, indicating that the twn2 mutant was homozygous for the kanamycin resistance (kanr) gene. However, the segregation ratio of kanr in the F2 generation was 486:428 (resistant/sensitive), which deviated significantly from the expected 3:1 ratio for a single dominant trait. Reciprocal crosses between a twn2 heterozygote and wild type showed that kanr was underrepresented in F1 progeny from both crosses (Table 1). Thus, the fitness of both gametes was decreased by the T-DNA. The twn2 mutation reduced the fitness of female gametes to a greater extent than the male gametes. In conjunction with the reduced viability of homozygous seeds, these observations seem adequate to account for the skewed F2 segregation ratio of the twn2 phenotype and the kanr trait.
Table 1.
Reciprocal crosses between twn2 heterozygotes and wild type
| Cross | Phenotype of F1 progeny
|
|
|---|---|---|
| kanr (%) | kans (%) | |
| ♀+/+ × ♂twn2/+ | 197 (45) | 243 (55) |
| ♀twn2/+ × ♂+/+ | 158 (34) | 309 (66) |
The expected ratio of kanr and kans is expected to be 1:1 if the T-DNA insertion does not affect gametogenesis.
To test whether the twn2 mutation was tagged by T-DNA, cosegregation of the twin embryo phenotype and kanr was analyzed in F3 families obtained by selfing F2 plants. All 80 F3 families derived from kanr F2 individuals produced twins at the expected frequency, while none of 50 families of kanamycin sensitivity (kans) F3 did. Therefore, we concluded that the twn2 mutant was probably tagged by T-DNA.
Reciprocal crosses between the twn2 mutant and the twn1 mutant (15) resulted in phenotypically wild-type F1 progeny. Hence, twn2 defines a new locus. In an attempt to obtain additional alleles of twn2, we screened 8,000 seeds from an ethyl methanesulfonate-mutagenized M2 population, and 18,000 seeds from a γ-irradiated M2 population on agar plates for seeds containing twin embryos. We identified eight additional mutants that produced twin seedlings. The new mutants represented at least two additional loci, designated twn3 and twn4. However, no new alleles of twn2 were isolated.
The twn2 Mutation Arrests Apical Cell Development.
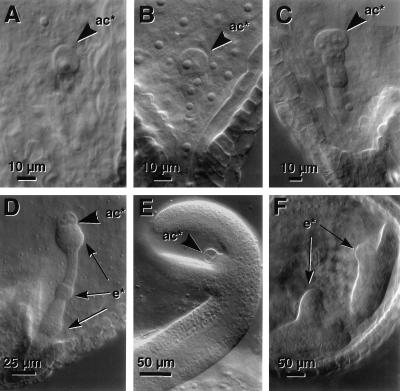
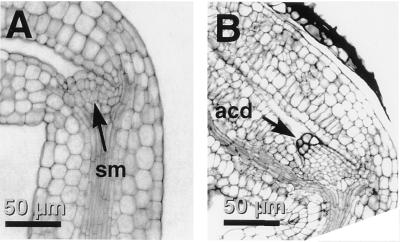
Microscopic comparison of the development of wild-type embryos (Fig. 2) with that of twn2 embryos (Fig. 3) revealed that the mutant exhibited a defect at an early stage of embryogenesis. In wild-type plants, within a few hours of fertilization, the zygote undergoes an asymmetric division, which gives rise to a smaller apical cell and a larger basal cell (Fig. 2A). The first division of the apical cell is longitudinal, whereas the basal cell divides transversely (Fig. 2B). Two more divisions by the daughters of the apical cell produces the octant stage embryo-proper that is only slightly larger than the apical cell (Fig. 2C). The embryo proper gives rise to all of the embryo except for its very base, the hypophysis and its derivatives, which is derived from the basal cell (Fig. 2 D–F). In twn2, the first division of the zygote is indistinguishable from that of wild type. However, after the first or second longitudinal division, the twn2 apical cell daughters cease further division and enlarge (Fig. 3A). This characteristic arrest and enlargement of the apical cells was observed in all of the several hundred embryos from homozygous twn2 plants that were examined. The daughters of the basal cell, which normally become the suspensor and hypophysis, undergo a series of divisions (Fig. 3 B and C), and eventually differentiate into one or more embryos (Fig. 3 D and F). The daughters of the apical cell remain in their original position throughout embryo development (Fig. 3 A–E) and can be seen even after germination (Fig. 4). They are distinguishable from the basal cell-derived embryo due to their somewhat darker stain with analine blue (Fig. 4).
Figure 2.
Wild-type embryo development. (A) Two cell stage. (B) Octant-dermatogen stage. (C) Early globular stage. (D) Mid-heart stage. (E) Early torpedo stage. (F) Late torpedo stage. ac, Apical cell; bc, basal cell; ec, embryonic cells; bd, derivatives of basal cell; ep, embryo proper; s, suspensor; co, cotyledon primordia or cotyledon; dh, derivatives of hypophysis; hy, hypocotyl.
Figure 3.
twn2 embryo development. (A) Two cell stage. (B) The apical cell appears to have arrested and the proximal basal cells have begun to swell. (C) Apical cells are arrested at the two to four cell stage and basal cells have begun to proliferate with out-of-file divisions. (D) Apical cell arrested and three putative embryonic cell masses have formed from proliferation of basal cells. (E) Late torpedo stage embryo showing persistence of arrested apical cells. (F) Malformed early torpedo stage duplicated embryos. ac*, Arrested apical cell; e*, ectopic embryos.
Figure 4.
Wild-type and twn2 shoot apical meristems. (A) Wild type. (B) twn2. sm, Shoot apical meristem; acd, arrested apical cell derivatives.
Because the seedling phenotype of the mutant was highly variable, we examined the phenotypes of several hundred embryos from the twn2 mutant at different stages. Arrest of the apical cells was observed in all embryos. However, development of the suspensor was highly variable. Based on the size and appearance of the embryonic structures in developing seeds it appeared that, in many cases, three or more embryos formed almost simultaneously. Because we rarely recovered seeds with more than two viable embryos it appeared that seeds containing more than two embryos generally failed to produce viable seedlings or that some of the embryos fail to develop fully when the number exceeds two. In other cases, there were large discrepancies in the stage of development of the multiple embryos. We believe that in these cases only a single viable embryo was produced or the secondary embryos produced only part of the overall embryo body (e.g., Fig. 1B). Thus, the incomplete penetrance and variability of the phenotype appeared to be due to the apparently stochastic initiation of embryo development from the basal cell lineage.
Molecular Basis of the twn2 Phenotype.
Southern blot analyses of genomic DNA from the twn2 mutant probed with the borders of the T-DNA region (20) indicated that it had four tandem copies of T-DNA (data not shown). Clones containing the left borders of the four copies of the T-DNA and their flanking regions were isolated from a genomic library of the mutant. Sequencing of the left borders and flanking regions of the four clones indicated that the four T-DNA copies formed one large cluster of ≈70 kb, consisting of both head–head and head–tail repeats. Two of the T-DNA borders were joined to plant DNA (data not shown). The two plant flanking regions hybridized to the same fragment on a Southern blot of wild-type genomic DNA, indicating that there were no major rearrangements in the mutant around the T-DNA insertion site (data not shown). The nucleotide sequence of the ≈4.5-kb region of the wild-type genome surrounding the insertion site was determined (GenBank accession number U93308). In addition, using the flanking regions of plant DNA as probes, 16 cDNA clones were isolated from two Arabidopsis cDNA libraries. These clones were found to represent two genes, a putative valyl tRNA synthetase (valRS), and a gene which we named decoy that encodes a 19,105-Da protein with low sequence homology to a yeast mitochondrial ribosomal protein (21). The longest clone of the valRS gene, present in plasmid pVRS19, was 3.6 kb in length and encoded a 973-amino acid polypeptide of 109,871 Da. The amino acid sequence of the valRS protein exhibited up to 47% sequence identity to valyl-tRNA synthetases from representatives of other orders (Table 2). The sequences of putative full-length cDNAs for both genes were deposited in GenBank as accession numbers U87586 and U89986, respectively.
Table 2.
Percent similarity (upper diagonal) and identity (lower diagonal) of Arabidopsis valRS amino acid sequence compared to valRS from other species
| valRS | M.j. | H.i. | E.c. | S.c. | H.s. | |
|---|---|---|---|---|---|---|
| valRS | 100 | 52 | 61 | 61 | 63 | 62 |
| Methanococcus jannaschii (M.j.) | 31 | 100 | 56 | 53 | 52 | 53 |
| Haemophilus influenza (H.i.) | 42 | 34 | 100 | 84 | 62 | 62 |
| Escherichia coli (E.c.) | 42 | 31 | 71 | 100 | 61 | 61 |
| Saccharomyces cerevisiae (S.c.) | 47 | 31 | 43 | 42 | 100 | 64 |
| Homo sapiens (H.s) | 45 | 29 | 42 | 41 | 47 | 100 |
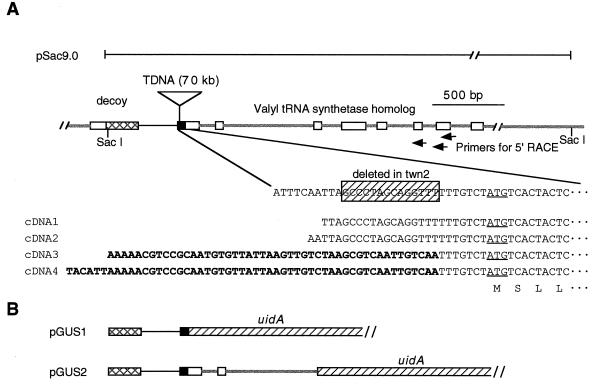
As shown in Fig. 5A, the 3′ end of the transcribed region of the decoy gene is ≈250 bp from the T-DNA insertion. The possible involvement of decoy in the twn2 phenotype was excluded for two reasons. First, northern blots of RNA from reproductive tissues, roots, and leaves probed with decoy showed that the twn2 mutation did not lower decoy expression (data not shown). In addition, transformation of the mutant with a genomic clone containing the decoy open reading frame and 2 kb of 5′ DNA failed to complement the mutant (data not shown).
Figure 5.
(A) Schematic drawing of the region surrounding the T-DNA insert in the twn2 mutant and some of the clones described herein. The T-DNA insert is not shown to scale. Exons are represented by boxes. The nontranslated 5′ region of the valRS gene is shown in black. The nontranslated 3′ region of the decoy gene is crosshatched. The coding region of the β-glucuronidase gene in constructs pGUS1 and pGUS2 are hatched. cDNA1 and cDNA2 represent two of the 5′ nucleotide sequences obtained by 5′ RACE of wild-type mRNA. cDNA3 an cDNA4 were obtained by 5′ RACE of mRNA from leaves of the twn2 mutant. Bold sequences represent sequences that correspond to T-DNA. (B) β-Glucuronidase constructs used in this study.
The approximate transcriptional start site of the wild-type valRS gene was determined by sequencing the 5′ ends of seven cDNA clones from the wild type. This analysis revealed that the insertion site of the T-DNA in the twn2 mutant was in the 5′ untranslated region of the valRS gene (Fig. 5A). As a result of the insertion event, 14 bp of genomic DNA at the insertion site was deleted (Fig. 5A). Northern blots probed with the valRS gene indicated that expression of the valRS gene in reproductive tissues was strongly depressed in the mutant (Fig. 6), suggesting that the mutation in the valRS gene might be responsible for the twn2 phenotype.
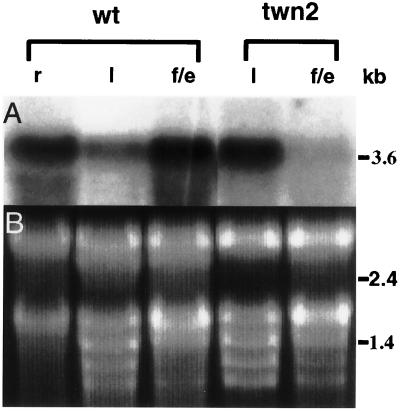
Figure 6.
TWN2 expression in wild-type and the twn2 mutant. (A) RNA blot probed with the 3.6 ApaI–SacI fragment from pVRS19. (B) Ethidium bromide-stained RNA gel. r, Root; l, leaf; f/e, flower and embryos.
To complement the mutant, a 9-kb SacI genomic fragment containing the valRS coding sequence along with 0.5 kb of 5′ flanking DNA was cloned into plant transformation vector pBIN-HYG (16) to produce plasmid pSac9.0. Because the 3′ end of the decoy gene is located only about 250 bp upstream of the transcriptional start site of the valRS gene, the fragment in pSAC9.0 contains part of the last exon of the decoy gene. This plasmid was used to transform homozygous twn2 plants. Transformants were selected on Murashige and Skoog medium containing both kanamycin and hygromycin. Because of the low fertility of the twn2 mutant, only one transformant was obtained. Both root and embryo development of this transformant was indistinguishable from the wild type and no hygromycin- and kanamycin-resistant twn mutants were recoved from among the progeny of this transformant.
To confirm the complementation results, twn2/+ plants were also transformed with pSac9.0. Seven primary transformants were obtained that were resistant to both kanamycin and hygromycin. From each of the primary transformants, 10 F2 plants were grown to maturity and their seeds (F3) were tested for hygromycin and kanamycin resistance. In each case, several plants with the genotype kanr/kanr hygr/hygs showed wild-type root and embryo development (data not shown). Therefore, we concluded that the twn2 mutant phenotype is caused by the inactivation of the valRS gene in the embryo by the T-DNA.
The valRS gene has been mapped on yeast artificial chromosomes yUP7G3, yUP24A3, yUP14C6, and yUP16B6, which are located on the top arm of chromosome 1 between DIS1 and EMB30. There does not appear to be any other closely related valRS gene in the Arabidopsis genome based on low stringency Southern analysis and PCR using degenerate primers to the conserved regions (data not shown). This may indicate that the valRS gene encodes both the cytoplasmic and mitochondrial forms of valyl-tRNA synthetase as has been previously reported for the alanyl-tRNA synthetase from Arabidopsis (22).
Expression of TWN2 in Wild-Type and twn2 Mutant.
To determine how a 70-kb insertion in an apparent housekeeping gene could cause the specific developmental phenotype in twn2, we examined the effect of the T-DNA insertion on the expression of the valRS gene. Total RNA from roots, leaves, and reproductive tissues (including flowers and developing seeds at various stages) from wild-type plants, and total RNA from leaves and reproductive tissues of twn2 plants were probed with the valRS cDNA. It was not possible to obtain enough root material from the twn2 mutant for a RNA blot. The results, shown in Fig. 6, indicated that the valRS gene was expressed in all three organs in wild-type plants, and as expected, the amount of steady-state mRNA in twn2 reproductive tissues is significantly reduced (Fig. 6). Unexpectedly, the amount of valRS mRNA in leaves of the mutant was higher than in the wild type (Fig. 6).
To determine where the valRS transcripts originated in the mutant, the 5′ ends of the valRS mRNA from leaves of the twn2 mutant were cloned by the 5′ RACE method. Two of the four cDNA clones sequenced contained sequences from the T-DNA (Fig. 5) indicating that the transcription start site was within the T-DNA. This observation raised the possibility that transcription of the gene in the mutant was initiated from leaf-specific promoter elements in the T-DNA. Alternatively, because of the small distance between the end of the decoy gene and the beginning of the valRS gene, we considered it possible that the expression of the gene in leaves might be activated by enhancer element(s) downstream of the T-DNA insertion site (e.g., in the introns of the valRS gene).
To test if 5′ introns were involved in the regulation of the gene in leaves, we constructed two valRS promoter fusions to the uidA (β-glucuronidase) reporter gene, shown in Fig. 5B. One construct, designated pGUS1, contained only the 5′ flanking and untranslated region and a piece of the last exon of the decoy gene, while the second (pGUS2) also contained the first two introns of the valRS gene. These constructs were transformed into Arabidopsis plants, and the level of β-glucuronidase activity in leaves and reproductive tissues was measured. The average leaf β-glucuronidase specific activity from 12 independent transformants containing the intronless construct and the intron-containing construct was 0.01 and 21.97 units, respectively. Similarly, in reproductive tissues, the levels of activity were 0.4 and 38.88 units, respectively (units of β-glucuronidase activity are pmol/min per μg of protein). Thus, the results of these experiments indicated that the presence of the first two introns increased the expression of the reporter gene by several hundred-fold. This indicates that the 5′ flanking sequence of the valRS gene does not function as a promoter in the absence of one or both of the first two introns, or that the introns greatly increase the stability of the mRNA. In view of the evidence showing that the valRS gene is transcribed at high rates when the 5′ flanking sequence is replaced by T-DNA in the twn2 mutant, we conclude that the first two introns contain an enhancer that can initiate transcription from various 5′ flanking sequences, including the T-DNA border.
DISCUSSION
The twn2 Phenotype Is Caused by Altered Expression of the valRS Gene.
The twn2 mutant is characterized by a high frequency of seeds containing fully or partially duplicated embryos. The viable seedlings give rise to fertile plants that are relatively normal in appearance but have short primary roots and reduced vigor. The mutant phenotype is caused by the insertion of ≈70 kb of T-DNA in the 5′ untranslated region of a putative valRS. The high degree of sequence identity between the Arabidopsis valRS gene and valyl-tRNA synthetases from other species leaves little doubt as to the function of the gene product. The T-DNA insertion reduces the level of steady-state mRNA for the gene in reproductive tissues, but not in leaves. Thus, the insertion mutation causes a tissue-specific reduction in the level of expression of the valRS gene. Because, the twn2-1 mutation represents a unique alteration of the 5′ flanking DNA sequence of the valRS gene, we do not consider it feasible to isolate additional mutant alleles that would exhibit the same pattern of altered gene expression. Also, because valyl-tRNA synthetase activity is an indispensable function, we do not consider it feasible to isolate null mutant alleles of the valRS gene. Thus, the following interpretation is offered with all the caveats associated with the use of single alleles and large DNA insertion–mutations.
Because it is not possible to obtain enough material to reliably measure expression of the valRS gene in apical and basal cells, we are unable to measure the relative levels of expression of the valRS gene in these cells of the mutant. However, in view of the fact that the insertion mutation caused differential accumulation of valRS mRNA in leaves and reproductive tissues, it is possible that the mutant gene is expressed at different levels in the apical cell and the basal cell lineage. Thus, we hypothesize that development of the apical cell arrests because of a lack of charged valyl-tRNA whereas the level of expression of the valRS gene in the basal cell is sufficiently high to support the growth requirements of this cell and its descendants. Alternatively, it is possible that the expression of the valRS gene is reduced to the same extent in the apical and basal cells but that reduced expression of the valRS gene is more detrimental to the development of the apical cell than to the basal cell. For instance, perhaps the reduced level of expression of the valRS gene disrupts critical early timing events in the apical cell that are not utilized in the basal cell.
Results from promoter–reporter fusions indicate that leaf-specific enhancer element(s) in the first two exons or introns have a major stimulatory effect on the expression of the gene in both leaves and reproductive tissues. This represents another of the many known instances in which introns have been shown to be important for the expression of genes (23–29). Sequencing of the 5′ ends of valRS transcripts from the mutant showed that some or all of the transcripts originate from a transcription start site in the T-DNA. We are not aware of any previous reports of transcripts originating in the T-DNA borders and consider this as evidence that the enhancer present in the first two introns of the valRS gene has relatively nonspecific requirements for an upstream sequence that can serve as an initiation site for transcription. However, the observation that the amount of mRNA for the valRS gene is reduced in developing seeds and flowers of the mutant, but is relatively normal in leaves, indicates that the activity of the putative enhancer is strongly modulated by tissue-specific factors. It is this fortuitous tissue specificity that is ultimately responsible for the twn2 mutant phenotype.
Cell Fate Determination in Arabidopsis.
Based on the properties of the twn1 mutant of Arabidopsis, it has previously been proposed that the apical cell actively inhibits embryonic development of the basal cell during early embryogenesis in angiosperms (15). In principle, this hypothesis might be directly tested by selectively killing the apical cell by laser ablation or a related physical method (30, 31). However, because the two-celled embryo is shrouded in maternal tissues, it has not yet been possible to devise a physical method for such experimental manipulations with angiosperms. To some extent, the twn2 mutant provides the genetic equivalent of an ablation experiment inasmuch as it selectively arrests development of the apical cell.
The development of multiple embryos in the twn2 mutant provides direct support for the hypothesis that the apical cell actively suppresses embryo formation by the basal cell lineage (15). The simplest interpretation of the evidence presented here is that the apical cell normally produces a factor that moves from the apical cell to the basal cell and suppresses the embryonic developmental potential of the basal cell in the two-celled embryo. We hypothesize that in the twn2 mutant, a deficiency in charged valyl-tRNA prevents synthesis of adequate amounts of the factor. It seems likely that the twn1 mutation (15), and the other twn mutations that we isolated while searching for a second twn2 allele, affect components of the apical–basal cell signaling pathway.
The presence of polyembryonic structures (e.g., Fig. 3D) suggests that suspensor-derived embryos did not suppress each others development. Thus, it seems that the cell or cells that first initiate the suspensor-derived embryos either do not produce the apical cell factor or produce it too late to prevent the suspensor cells from entering the embryogenic state. However, the suspensor-derived embryos appeared to develop at relatively evenly spaced locations along the axis of the polyembryonic tissue mass. Thus, we speculate that once a group of cells begins to organize an embryonic structure, the incipient embryo produces or consumes a diffusable factor that prevents additional embryos from initiating nearby. The presence of duplicated organs on some embryos (e.g., Fig. 1B) may indicate that if two embryos simultaneously initiate nearby, they may share some of the factors that regulate pattern formation. Thus, for instance, we hypothesize that the seedling in Fig. 1B arose from two cells or groups of cells undergoing simultaneous embryonic initiations. We envision that at an early stage of pattern formation, the incipient apical region of one group of cells produced a signal that informed both groups of cells that a shoot apex was being formed from a subpopulation of cells that was in appropriate proximity to both groups of cells, and this suppressed formation of a second shoot.
The hypothetical involvement of intercellular signaling mechanisms proposed here is in apparent contrast to the situation in the brown algae Fucus. Embryogenesis of Fucus resembles higher plant embryogenesis in several ways, and it has been proposed that some mechanisms of Fucus embryo development may parallel that of higher plants (32, 33). In Fucus, the first asymmetric cell division gives rise to a rhizoid cell that develops into the basal root-like tissue (holdfast) and an apical thallus cell that produces the vegetative and reproductive shoot (34). However, cell fate determination in the two cell stage in Fucus does not appear to rely on communication with the neighboring cell. Rather, it has been proposed that the differentiated state of the cell is determined by the contact of the cell with the cell wall. For example, ablation of one cell in the two cell stage does not affect the fate of the remaining cell (30, 31). Moreover, contact of one cell type with the isolated cell wall of the other cell type caused its fate to be switched (31). The exact nature of the fate determination factor(s) is not known, but based on the phenotype of twn2, it is unlikely to be the same in higher plants.
Acknowledgments
We thank Colby Starker for technical assistance, David Meinke and George Haughn for criticism and advice, Dan Vernon and David Meinke for doing complementation tests with twn1, and Christoph Benning for sharing mutant lines. This work was supported in part by a grant from the U.S. Department of Energy (DE-FG02-94ER20133).
ABBREVIATIONS
- RACE
rapid amplification of cDNA ends
- kanr
kanamycin resistance
- kans
kanamycin sensitivity
- hygr
hygromycin resistance
- hygs
hygromycin sensitivity
Footnotes
References
- 1.Mansfield S G, Briarty L G, Erni S. Can J Bot. 1991;69:447–460. [Google Scholar]
- 2.Jensen W A. Planta. 1968;79:346–366. doi: 10.1007/BF00386917. [DOI] [PubMed] [Google Scholar]
- 3.Russell S D. Plant Cell. 1993;5:1349–1359. doi: 10.1105/tpc.5.10.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mansfield S G, Briarty L G. Can J Bot. 1991;69:461–476. [Google Scholar]
- 5.Juergens G, Mayer U. In: Embryos. Bard J, editor. London: Wolfe; 1994. pp. 7–22. [Google Scholar]
- 6.Meinke D W. In: Arabidopsis. Meyerowitz E M, Somerville C R, editors. Plainview, New York: Cold Spring Harbor Lab. Press; 1994. pp. 253–295. [Google Scholar]
- 7.Meinke D W. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:369–394. [Google Scholar]
- 8.Priess J R, Schnabel H, Schnabel R. Cell. 1987;51:601–611. doi: 10.1016/0092-8674(87)90129-2. [DOI] [PubMed] [Google Scholar]
- 9.Liu C M, Xu Z H, Chua N H. Plant Cell. 1993;5:621–630. doi: 10.1105/tpc.5.6.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C M, Xu Z H, Chua N H. Plant J. 1993;3:291–300. [Google Scholar]
- 11.Kirk M M, Ransick A, McRae S E, Kirk D L. J Cell Biol. 1993;123:191–208. doi: 10.1083/jcb.123.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gurdon J B. Cell. 1992;68:185–199. doi: 10.1016/0092-8674(92)90465-o. [DOI] [PubMed] [Google Scholar]
- 13.Yadegari R, De Paiva G R, Laux T, Koltunow A M, Apuya N, Zimmerman J L, Fischer R L, Harada J J, Goldberg R B. Plant Cell. 1994;6:1713–1729. doi: 10.1105/tpc.6.12.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz B W, Yeung E C, Meinke D W. Development (Cambridge, UK) 1994;120:3235–3245. doi: 10.1242/dev.120.11.3235. [DOI] [PubMed] [Google Scholar]
- 15.Vernon D M, Meinke D W. Dev Biol. 1994;165:566–573. doi: 10.1006/dbio.1994.1276. [DOI] [PubMed] [Google Scholar]
- 16.Feldman K. Plant J. 1991;1:71–82. [Google Scholar]
- 17.Becker D. Nucleic Acids Res. 1990;18:203. doi: 10.1093/nar/18.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katavic V, Haughn G W, Reed D, Martin M, Kunst L. Mol Gen Genet. 1994;245:363–370. doi: 10.1007/BF00290117. [DOI] [PubMed] [Google Scholar]
- 19.Danhof L R, Bariola P A, Green P J. Plant Physiol (Suppl) 1996;111S:164. (abstr.). [Google Scholar]
- 20.Zambryski P, Joos H, Genetello C, Leemans J, Van Montagu M, Schell J. EMBO J. 1983;2:2143–2150. doi: 10.1002/j.1460-2075.1983.tb01715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grohmann L, Graack H-R, Kruft V, Choli T, Goldschmidt-Reisin S, Kitakawa M. FEBS Lett. 1991;284:51–56. doi: 10.1016/0014-5793(91)80759-v. [DOI] [PubMed] [Google Scholar]
- 22.Mireau H, Lancelin D, Small I D. Plant Cell. 1996;8:1027–1039. doi: 10.1105/tpc.8.6.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bornstein P, Mckay J, Morishima J K, Devarayalu S, Gelinas R E. Proc Natl Acad Sci USA. 1987;84:8869–8873. doi: 10.1073/pnas.84.24.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Callis J, Fromm M, Walbot V. Genes Dev. 1987;1:1183–1200. doi: 10.1101/gad.1.10.1183. [DOI] [PubMed] [Google Scholar]
- 25.Cohen J B, Levinson A D. Nature (London) 1988;334:119–124. doi: 10.1038/334119a0. [DOI] [PubMed] [Google Scholar]
- 26.Fu H, Kim S Y, Park W D. Plant Cell. 1995;7:1387–1403. doi: 10.1105/tpc.7.9.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gillies S D, Morrison S L, Oi V T, Tonegawa S. Cell. 1983;33:717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- 28.Rossi P, de Crombrugghe B. Proc Natl Acad Sci USA. 1987;84:5590–5594. doi: 10.1073/pnas.84.16.5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tiffany H L, Handen J S, Rosenberg H F. J Biol Chem. 1996;271:12387–12393. doi: 10.1074/jbc.271.21.12387. [DOI] [PubMed] [Google Scholar]
- 30.Kropf D L, Coffman H R, Kloareg B, Glenn P, Allen V W. Dev Biol. 1993;160:303–314. doi: 10.1006/dbio.1993.1309. [DOI] [PubMed] [Google Scholar]
- 31.Berger F, Taylor A, Brownlee C. Science. 1994;263:1421–1423. doi: 10.1126/science.263.5152.1421. [DOI] [PubMed] [Google Scholar]
- 32.Berger F, Brownlee C. Biol Cell. 1995;84:7–11. [Google Scholar]
- 33.Quatrano R S, Shaw S L. Trends Plant Sci. 1997;2:15–21. [Google Scholar]
- 34.Kropf D L. Microbiol Rev. 1992;56:316–339. doi: 10.1128/mr.56.2.316-339.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]