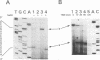Abstract
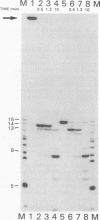
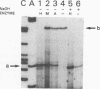
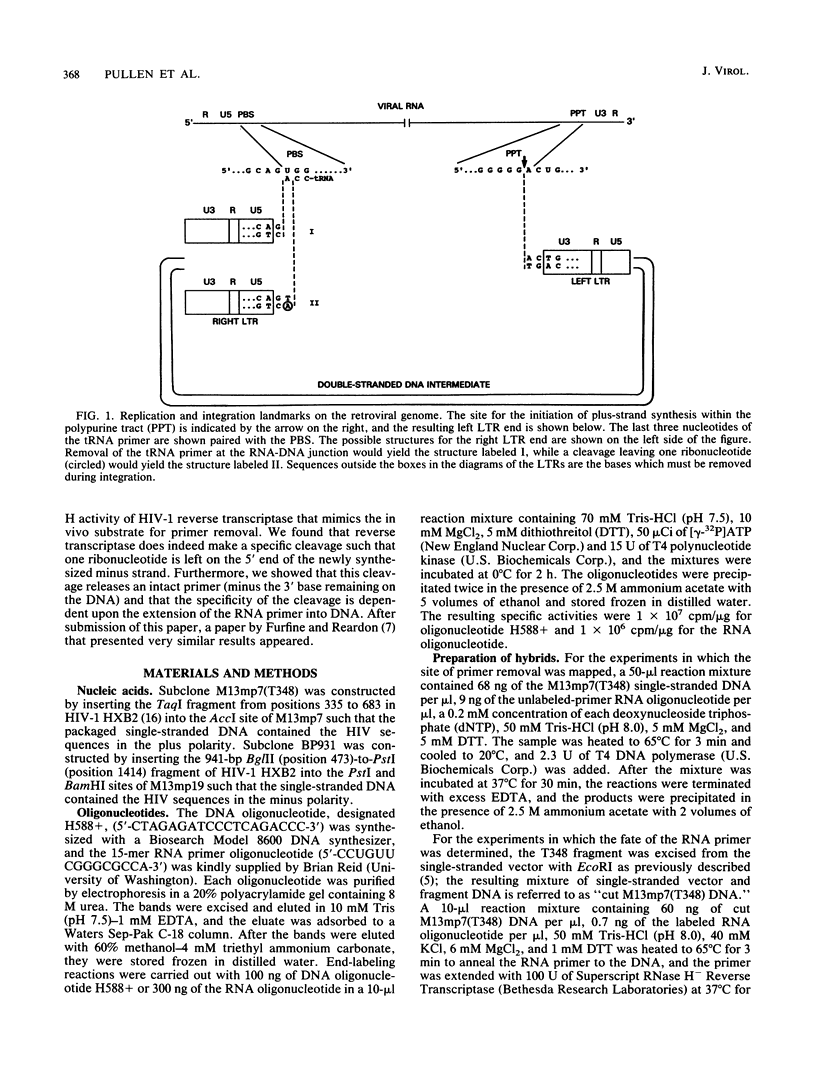
A synthetic RNA oligonucleotide (15-mer) corresponding to the 3' end of the lysine tRNA primer was hybridized to single-stranded DNA containing the human immunodeficiency virus type 1 (HIV-1) primer-binding site and extended with a DNA polymerase. The resulting structures were used to study primer removal by the RNase H activity of HIV-1 reverse transcriptase. The initial cleavage event removes the RNA primer as a 14-mer and leaves a single ribonucleotide A residue bound to the 5' end of the DNA strand. This result explains the observation by several groups that HIV-1 circle junctions contain 4 bp that are not present in the integrated provirus instead of the predicted 3 bp. Subsequent cleavage events occur at other sites internal to the RNA molecule, and the ribonucleotide A residue on the end of the DNA strand is ultimately removed. Therefore, the biologically relevant cleavage that produces the 14-mer reflects the kinetics of the reaction as well as a specificity for nucleic acid sequence. When the RNA oligonucleotide alone was hybridized to the primer-binding site and tested as a substrate for HIV-1 RNase H, the cleavage pattern near the 3' end of the RNA was altered.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown P. O., Bowerman B., Varmus H. E., Bishop J. M. Correct integration of retroviral DNA in vitro. Cell. 1987 May 8;49(3):347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- Champoux J. J., Gilboa E., Baltimore D. Mechanism of RNA primer removal by the RNase H activity of avian myeloblastosis virus reverse transcriptase. J Virol. 1984 Mar;49(3):686–691. doi: 10.1128/jvi.49.3.686-691.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colicelli J., Goff S. P. Sequence and spacing requirements of a retrovirus integration site. J Mol Biol. 1988 Jan 5;199(1):47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- Ellis J., Bernstein A. Retrovirus vectors containing an internal attachment site: evidence that circles are not intermediates to murine retrovirus integration. J Virol. 1989 Jun;63(6):2844–2846. doi: 10.1128/jvi.63.6.2844-2846.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finston W. I., Champoux J. J. RNA-primed initiation of Moloney murine leukemia virus plus strands by reverse transcriptase in vitro. J Virol. 1984 Jul;51(1):26–33. doi: 10.1128/jvi.51.1.26-33.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini G., Gurgo C., Guo H. G., Gallo R. C., Collalti E., Fargnoli K. A., Hall L. F., Wong-Staal F., Reitz M. S., Jr Sequence of simian immunodeficiency virus and its relationship to the human immunodeficiency viruses. Nature. 1987 Aug 6;328(6130):539–543. doi: 10.1038/328539a0. [DOI] [PubMed] [Google Scholar]
- Furfine E. S., Reardon J. E. Human immunodeficiency virus reverse transcriptase ribonuclease H: specificity of tRNA(Lys3)-primer excision. Biochemistry. 1991 Jul 23;30(29):7041–7046. doi: 10.1021/bi00243a001. [DOI] [PubMed] [Google Scholar]
- Gilboa E., Mitra S. W., Goff S., Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979 Sep;18(1):93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- Huber H. E., Richardson C. C. Processing of the primer for plus strand DNA synthesis by human immunodeficiency virus 1 reverse transcriptase. J Biol Chem. 1990 Jun 25;265(18):10565–10573. [PubMed] [Google Scholar]
- Kulkosky J., Katz R. A., Skalka A. M. Terminal nucleotides of the preintegrative linear form of HIV-1 DNA deduced from the sequence of circular DNA junctions. J Acquir Immune Defic Syndr. 1990;3(9):852–858. [PubMed] [Google Scholar]
- Lobel L. I., Murphy J. E., Goff S. P. The palindromic LTR-LTR junction of Moloney murine leukemia virus is not an efficient substrate for proviral integration. J Virol. 1989 Jun;63(6):2629–2637. doi: 10.1128/jvi.63.6.2629-2637.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S. W., Chow M., Champoux J., Baltimore D. Synthesis of murine leukemia virus plus strong stop DNA initiates at a unique site. J Biol Chem. 1982 Jun 10;257(11):5983–5986. [PubMed] [Google Scholar]
- Omer C. A., Faras A. J. Mechanism of release of the avian rotavirus tRNATrp primer molecule from viral DNA by ribonuclease H during reverse transcription. Cell. 1982 Oct;30(3):797–805. doi: 10.1016/0092-8674(82)90284-7. [DOI] [PubMed] [Google Scholar]
- Omer C. A., Resnick R., Faras A. J. Evidence for involvement of an RNA primer in initiation of strong-stop plus DNA synthesis during reverse transcription in vitro. J Virol. 1984 May;50(2):465–470. doi: 10.1128/jvi.50.2.465-470.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullen K. A., Champoux J. J. Plus-strand origin for human immunodeficiency virus type 1: implications for integration. J Virol. 1990 Dec;64(12):6274–6277. doi: 10.1128/jvi.64.12.6274-6277.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner L., Haseltine W., Patarca R., Livak K. J., Starcich B., Josephs S. F., Doran E. R., Rafalski J. A., Whitehorn E. A., Baumeister K. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985 Jan 24;313(6000):277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- Rattray A. J., Champoux J. J. The role of Moloney murine leukemia virus RNase H activity in the formation of plus-strand primers. J Virol. 1987 Sep;61(9):2843–2851. doi: 10.1128/jvi.61.9.2843-2851.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick R., Omer C. A., Faras A. J. Involvement of retrovirus reverse transcriptase-associated RNase H in the initiation of strong-stop (+) DNA synthesis and the generation of the long terminal repeat. J Virol. 1984 Sep;51(3):813–821. doi: 10.1128/jvi.51.3.813-821.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank P. R., Varmus H. E. Virus-specific DNA in the cytoplasm of avian sarcoma virus-infected cells is a precursor to covalently closed circular viral DNA in the nucleus. J Virol. 1978 Jan;25(1):104–104. doi: 10.1128/jvi.25.1.104-104.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C., Goff S., Gilboa E., Paskind M., Mitra S. W., Baltimore D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: implications for retrovirus integration. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3932–3936. doi: 10.1073/pnas.77.7.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. S., Kim S. Y., Roth M. J. Analysis of long terminal repeat circle junctions of human immunodeficiency virus type 1. J Virol. 1990 Dec;64(12):6286–6290. doi: 10.1128/jvi.64.12.6286-6290.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorge J., Hughes S. H. Polypurine tract adjacent to the U3 region of the Rous sarcoma virus genome provides a cis-acting function. J Virol. 1982 Aug;43(2):482–488. doi: 10.1128/jvi.43.2.482-488.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J., Sharmeen L. Retrovirus genome replication: priming specificities of plus-strand DNA synthesis. J Cell Sci Suppl. 1987;7:189–195. doi: 10.1242/jcs.1987.supplement_7.14. [DOI] [PubMed] [Google Scholar]
- Whitcomb J. M., Kumar R., Hughes S. H. Sequence of the circle junction of human immunodeficiency virus type 1: implications for reverse transcription and integration. J Virol. 1990 Oct;64(10):4903–4906. doi: 10.1128/jvi.64.10.4903-4906.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagury J. F., Franchini G., Reitz M., Collalti E., Starcich B., Hall L., Fargnoli K., Jagodzinski L., Guo H. G., Laure F. Genetic variability between isolates of human immunodeficiency virus (HIV) type 2 is comparable to the variability among HIV type 1. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5941–5945. doi: 10.1073/pnas.85.16.5941. [DOI] [PMC free article] [PubMed] [Google Scholar]