Abstract
Mutant forms of the BRCA2 gene contribute significantly to hereditary breast cancer. Isolation of the normal and mutant forms of the BRCA2 gene with its natural promoter would greatly facilitate analysis of the gene and its contribution to breast cancer. We have accomplished the direct isolation of the 90-kb gene from total human DNA by transformation-associated recombination in yeast using a small amount of 5′ and 3′ BRCA2 sequence information. Because the entire isolation procedure of a single chromosomal gene could be accomplished in approximately 2 weeks, the transformation-associated recombination cloning approach is readily applicable to studies of chromosome alterations and human genetic diseases.
As much as 10% of female breast cancers are considered hereditary, and approximately one-third are associated with the BRCA2 gene (1–3). BRCA2 mutations also confer susceptibility to male breast cancer (summarized in ref. 4). Studies of BRCA2-based cancers would be greatly facilitated by cloning of normal and mutated genomic copies from an individual. However, the isolation of the genomic copy of this gene, or for that matter any human gene identified with a disease, as an artificial chromosome in yeast (a YAC) or Escherichia coli [as a bacterial artificial chromosome (BAC)] is a long and labor-intensive process.
Previously we demonstrated that transformation-associated recombination (TAR) in the yeast Saccharomyces cerevisiae can be used to isolate random fragments of human DNA from rodent/human monochromosomal and radiation hybrid cell lines as linear or circular YACs (5, 6). The TAR cloning method as applied to human DNA is based on co-penetration into yeast spheroplasts of the DNA along with the vector(s) DNA that contains a repeat common to the human DNA, followed by recombination between the vector(s) and the human DNA to establish a YAC. Propagation of the YAC can occur if the human DNA contains sequences that can function as autonomously replicating sequences (ARS) in yeast (these are common in human DNA with approximately one ARS-like sequence per 20 to 40 kb (ref. 5 and references therein; ref. 7). More than a thousandfold enrichment for human DNA was achieved when a commonly occurring human repeat Alu was included in the TAR cloning vector (5, 6). Based on these results we investigated whether a specific single-copy gene could be isolated directly from total human DNA. We chose the BRCA2 gene for our study because of its breast cancer relevance and the fact that the entire ≈90-kb gene has not been isolated, although fragments have been identified in various artificial chromosome libraries (8, 9).
MATERIALS AND METHODS
Yeast Strain and Transformation.
For transformations, the highly transformable S. cerevisiae strain VL6–48 (MATa, his3-Δ200, trp1-Δ1, ura3–52, lys2, ade2–101, met14) (10) that has HIS3 deleted was used. Spheroplasts that enable efficient transformation were generated using a previously described protocol (5). Agarose plugs (100 μl) containing approximately 5 μg of high molecular weight human DNA were prepared from normal human fibroblasts MRC-5 (American Type Culture Collection) and used for yeast transformation (5, 6). Linearized vector (1 μg) was added to the DNA-containing plugs before they were treated with agarase and presented to spheroplasts. Yeast transformants were selected on synthetic complete medium plates lacking histidine.
Construction of TAR Cloning Vector.
The TAR circularizing vector pVC-BC2, which contained 5′ and 3′ sequences of the human BRCA2 gene, was constructed from the vector pVC1 (Alu-CEN6-HIS3-TEL) that was previously used for selective cloning of human DNA from human/rodent hybrid cell lines (5, 6). The Alu and telomere TEL sequences in pVC1 were replaced by a 669-bp XhoI–EcoRI fragment corresponding to promoter region and a 308-bp EcoRI–BamHI fragment corresponding to 3′ sequence of BRCA2. Both BRCA2 targeting sequences were developed based on the information available from cDNA and partial genomic BRCA2 DNA sequences and lacked any human repeat elements. The 5′ targeting sequence of 669 bp corresponds to positions 47,654–48,322 in the genomic sequence (accession number Z74739), and the 3′ targeting sequence of 308 bp corresponds to the last exon of BRCA2 [positions 10,179–10,486 in the BRCA2 cDNA sequence (accession number U43746)]. The 5′ targeting sequence is approximately 7.0 kb upstream of the ATG initiation codon, and the 3′ BRCA2 sequence is located in exon 27 and contains the stop codon. Both BRCA2 targeting fragments (hooks) were checked for ARS activity. The 3′ fragment contained an ARS sequence based on a high transformation efficiency of the plasmid containing the fragment. An ARS consensus sequence was identified near the stop codon at positions 10,469–10,480 in the BRCA2 cDNA sequence. The sequence was mutagenized to AGA AGT ACA by PCR before the targeting sequence was cloned into the TAR vector pVC-BC2. The resulting sequence was shown to lack ARS activity. The targeting sequences were cloned into pVC1 plasmid in orientation corresponding to their orientation in the genome. The vector pVC-BC2 was cut with EcoRI (the site is located between the targeting sequences) before transformation to yield a molecule bounded by the BRCA2 sequences.
PCR Analysis.
Three pairs of 20-bp primers were used for PCR characterization of YAC pools: JAD18R and JAD18L, specific for internal exon 11 sequence; J3R and J3L, specific for 3′ sequence; and J5R and J5L, specific for promoter region of BRCA2. JAD18R and JAD18L amplify a 320-bp sequence of exon 11 (11); J3R (5′-TTCTGAACTGGTGGGAGCAG-3′) and J3L (5′-CCACCTGTTAGTCCCATTTG-3′) amplify a 291-bp sequence of the genomic copy of BRCA2. The J3R is in the 3′ targeting BRCA2 hook sequence on the vector (positions 10,340–10,359 in the BRCA2 cDNA sequence, accession number U43746), and J3L corresponds to a region upstream from the hook (7,857–7,876 in genomic sequence, accession number Z73359). The PCR product is diagnostic of recombination between the TAR vector and the 3′ region of the genomic BRCA2. The J5L (5′-CTACCCTTGAGGTTTCAAGG-3′) and J5R (5′-AGGGAGTTCTCAGAACTATG-3′) primers amplify a 435-bp sequence of the BRCA2 promoter region. The J5L sequence is located in the 5′ targeting hook 90 bp upstream of the end of the 5′ hook, and the J5R sequence is 345 bp beyond the 5′ end of the hook (positions 48,667–48,648 in genomic sequence, accession number Z74739). Because J5L is 344 bp downstream of the end of the 5′ hook, generation of a 434-bp PCR product is diagnostic of recombination of the 5′ targeting sequence of the TAR vector with the promoter sequence of genomic copy of BRCA2. The BRCA2 coding region in YAC clones was examined by PCR using 26 pairs of primers for exons 2–27 as previously described (11). Yeast genomic DNA isolated from transformants (1 μg) was amplified using primers under the following standard PCR conditions: 50 mM KCl, 10 mM Tris⋅HCl (pH 9.0), 3.0 mM MgCl2, and 0.2 mM dTTP, dCTP, dGTP, and dATP in a final volume of 50 μl. Thermocycling conditions consisted of 35 cycles of 1 min at 94°C, 45 s at 55°C, and 5 min at 68°C, followed by one cycle of 10-min extension at 72°C in a 9600 Thermocycler (Perkin–Elmer).
Characterization of YAC Clones.
Chromosomal-size DNAs from transformants were separated by transverse alternating field electrophoresis (TAFE), blotted, and hybridized with total human genomic DNA as previously described (5, 6). To estimate the size of circular YACs, agarose DNA plugs were exposed to a low dose of γ rays (5 Krad) before TAFE analysis. To identify fragments containing Alu sequences (Alu profiles), 1 μg of total yeast DNA was digested to completion with TaqI. Samples were run by gel electrophoresis, transferred to a nylon membrane, and hybridized with an Alu probe (6). The frequency of YAC loss was determined by measuring the loss of the centromere-linked HIS3 marker, as previously described for yeast transformants (6). In brief, colonies were streaked onto nonselective medium and the frequency of His− colonies was determined by replica plating onto selective synthetic medium that lacked histidine.
BRCA2 YAC Retrofitting.
A new yeast-bacteria-mammalian-cells shuttle vector, BRV1, was constructed for retrofitting of large, circular YACs to BACs containing the mammalian selectable marker NeoR (unpublished work). Recombination of BRV1 with a YAC in yeast leads to replacement of the ColE1 origin of replication in TAR cloning vector by a cassette containing the F factor origin replication, the chloramphenicol acetyltransferase (CmR) gene, the NeoR gene, and the URA3 yeast selectable marker. The retrofitted YACs were moved to E. coli by electroporation as described previously (6).
RESULTS
TAR Cloning of Human BRCA2 Gene.
The TAR cloning scheme for isolating the BRCA2 gene described in Fig. 1 utilizes cDNA and partial genomic sequence information. A centromere-based yeast TAR vector, pVC-BC2, was created so that when linearized, one end contained 669 bp of the BRCA2 promoter sequence approximately 7 kb from the start codon and the other end contained a 308-bp sequence of the last exon (including the stop codon; see Materials and Methods). To exclude transformants arising from vector religation, the BRCA2 fragments were checked for ARS activity. ARS activity of the 3′ targeting fragment was detected and inactivated by site-directed mutagenesis. Thus, propagation of the TAR vector then depended on acquisition of an ARS from the cloned material.
Figure 1.
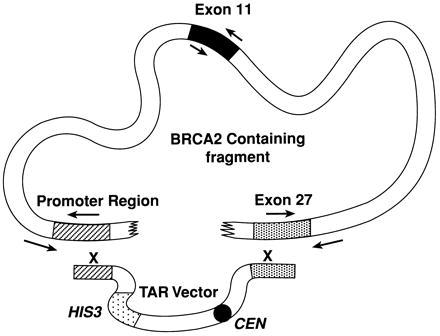
Isolation of the human BRCA2 gene as a circular YAC using the TAR cloning method. Yeast spheroplasts were transformed with genomic human DNA along with a TAR cloning vector containing promoter (striped box) and terminal exon 27 (dotted box) sequences of BRCA2 (see Materials and Methods) at the ends of the linearized plasmid. Recombination between the sequences in the vector and BRCA2 genomic DNA led to the establishment of a circular YAC because of ARS-like sequences in the BRCA2 gene. CEN corresponds to the yeast chromosome VI centromere, and HIS3 is a selectable marker. Arrows indicate the positions of primers used for initial identification of clones containing the BRCA2 gene.
Ten transformation experiments were carried out with freshly prepared yeast spheroplasts as previously described for the cloning of human DNA from a human/rodent hybrid cell line (5, 6), and approximately 1,000 colonies were obtained. (The yield of transformants per 5 μg of human DNA using 1 μg of vector and 5 × 108 spheroplasts varied between 20 and 150.) To identify BRCA2-containing transformants, 1,000 of them were combined into 33 pools and examined by PCR for the presence of unique BRCA2 sequences not present in the vector. Three pairs of primers were utilized that identified exon 11 of the BRCA2 gene and the junctions of the BRCA2 promoter and 3′ sequences adjacent to the sequences in the vector (Fig. 1). There were three pools in which PCR products were obtained for all three primer pairs. The PCR products corresponding to exon 11 and the junctions next to the 5′ and the 3′ targeting sequences in the vector were sequenced and found to match the expected BRCA2 sequence (data not shown). None of the pools yielded products with only one or two of the three primer pairs. The three putative BRCA2 genes corresponded to independent isolates, because the pools were combined from different transformation mixes. Individual clones containing the BRCA2 sequence were isolated from each pool (clones 27, 32, and 60) and used for further analysis.
Physical Analysis of YAC Clones Containing BRCA2 Gene.
Several approaches were taken to establish the integrity and stability of the cloned material in the three isolates. Although a complete genomic copy of BRCA2 had not been isolated previously, an analysis of information about the cDNA and overlapping partial genomic clones revealed that the entire genomic region between the sequences used for TAR cloning should be 90,442 bp (refs. 8 and 9 and references therein). As expected for a circular YAC (6, 12), the BRCA2 hybridizing material was retained in the starting wells of a TAFE gel used for separation of chromosome-size DNAs. A low dose of ionizing radiation (12) was used to induce an average of less than one random break in the circular molecules. The largest linear molecules, corresponding to a single break, were 100 kb (Fig. 2). Because the TAR vector was 7 kb, the detectable size of the cloned human material corresponded to that predicted if the entire BRCA2 gene had been cloned.
Figure 2.
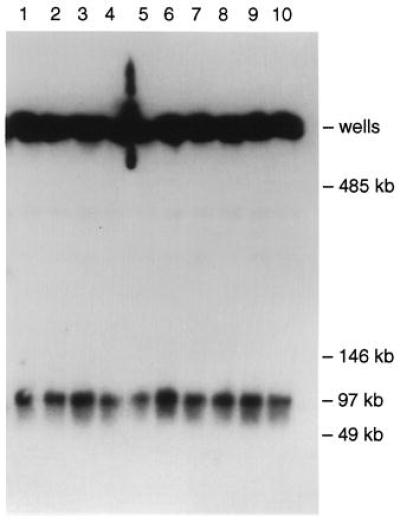
Characterization of circular BRCA2 YACs obtained by TAR cloning from total human DNA. Chromosomal-size DNA was isolated from transformants containing BRCA2 YACs, exposed to a low dose of γ rays (5 Krad), separated by TAFE gel electrophoresis, and blot hybridized with total human genomic DNA (6). The strong signals at the positions of the starting wells correspond to large circular molecules. The leading bands at approximately 97 kb correspond to molecules linearized by the radiation. Four subclones of isolates 27 and 32 (lanes 1–4 and 5–8, respectively) and two subclones of isolate 60 (lanes 9 and 10) were analyzed.
Alu profiles were determined for the three YACs and they were found to be identical. The three isolates were outgrown for more than 80 generations, and single-colony isolates were subsequently obtained and examined for YAC size and Alu profiles. Among 11 subclones obtained from the 3 original isolates, all were the expected size and exhibited identical Alu profiles, suggesting that the cloned sequence was stable during propagation (Fig. 3). Similar to previous observations with circular YACs generated by TAR cloning (6), the BRCA2 YACs were accurately segregated. The loss rate of BRCA2 YACs was 2–3 × 10−3 per cell per generation.
Figure 3.
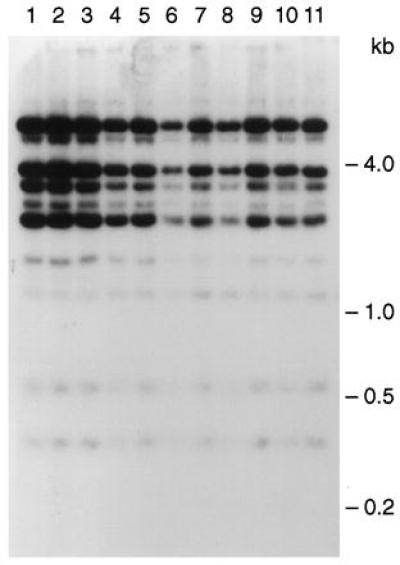
Alu-profile characterization of YACs containing BRCA2. Subclones of the three initial isolates were characterized and compared in terms of Alu profiles of restriction fragments. Total yeast DNA was isolated from each subclone and digested to completion with TaqI. Fragments were separated by gel electrophoresis, transferred to a nylon membrane, and hybridized with an Alu probe. Four subclones of isolates 27 (lanes 1–4) and 32 (lanes 5–8) and three subclones of isolate 60 (lanes 9–11) were analyzed.
To establish the integrity of the BRCA2 clones, the YACs were analyzed by PCR using 26 pairs of primers corresponding to exons 2–27 of BRCA2 (11). The expected sizes of PCR products was obtained with each primer pair, and these matched the PCR products from total genomic DNA (Fig. 4); no bands were observed using only yeast DNA (data not shown). (Presented in Fig. 4 are results with three exons and two junction primer pairs; comparable results were obtained with the rest of the exons.) Finally, we examined the restriction pattern of the YACs. Analysis of the expected cloned genomic sequence of BRCA2 revealed four MluI sites (refs. 8 and 9 and references therein); there were none in the pVC-BC2 vector. The expected four fragments were identified in a MluI digest of total genomic DNA isolated from transformants containing the BRCA2 YACs by probing with the Alu consensus sequence (data not shown). All three BRCA2 YACs were moved from yeast to E. coli cells using a BAC/NeoR retrofitting vector, BRV1, similar to that described in ref. 13. BRCA2 BAC/YAC/NeoR derivatives were analyzed by three restriction endonuclease digestions (MluI, BamHI, and MluI + BamHI). No differences between BRCA2 clones were found, and their restriction patterns corresponded to the published sequence of BRCA2 (data not shown).
Figure 4.
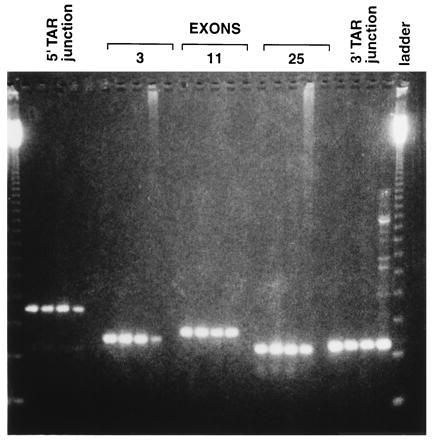
PCR analysis of YACs containing BRCA2. The presence of BRCA2 coding region in the YAC clones was examined by PCR using 26 pairs of exon primers and 2 pairs of junction primers. Presented in this figure are PCR products obtained for exons 3, 11, and 25 and for the 5′ and 3′ TAR vector junction sequences. (We note that comparable results were obtained for the other primers, in that the PCR products with the YACs matched those of the total human DNA.) The first three lanes for each set of PCR primers correspond to products obtained from the three independent BRCA2 YAC isolates. The fourth lane corresponds to the product generated from total human DNA. (In the PCR analysis of the 3′ TAR junction some additional bands were observed with the total human DNA that were due to PCR artifacts.) The fifth lane was a control in which no DNA was added (there was no fifth lane for the 3′ TAR junction). The first and last lanes contained 123-bp ladders.
We conclude from these studies that a single-copy BRCA2 gene can be directly and accurately isolated from genomic DNA by TAR cloning.
DISCUSSION
We have described the selective isolation of a single-copy gene from total human DNA by TAR cloning. Our success in the direct isolation of the BRCA2 single-copy gene was due, in part, to the use of a TAR vector lacking a yeast origin of replication, thereby reducing the background of transformants that might be due to vector religation. Previously we showed that the presence of an ARS in a TAR vector resulted in only 30% of the clones containing human DNA (5). Similarly, a high-transformation background was observed during the recombinational cloning of a 30-kb adenovirus DNA from excess of mouse DNA by two ARS-containing vectors carrying sequences homologous to virus (14). In the present experiments, the relatively low background obtained with the BRCA2 vector appeared to result from illegitimate recombination with human DNA, because most (greater than 80%) of the transformants contained circular YACs that hybridized with human DNA (data not shown). We note that the approach described here has also been applied to the isolation of two other single-copy genes from total human DNA (BRCA1 and HPRT; unpublished data).
Physical analysis of BRCA2 YACs are consistent with a high fidelity of gene isolation by TAR. Nevertheless, additional experiments are required to exclude possible small changes in cloned material such as point mutations or small deletions. The ability to isolate the entire human BRCA2 gene will benefit further studies into the function of this gene as well as aid in understanding the etiology of BRCA2-associated cancers. Little is known about the function of this gene. The opportunity to isolate and transfer the entire gene to tumor-derived human cell lines in which the gene is nonfunctional (15) will enable direct investigation of the consequences of the gene under the control of its natural promoter. Once its function is determined, mutant forms of BRCA2 can be rapidly isolated by TAR cloning from DNA of breast cancer families and tested for function. Because DNA transfection often leads to the incorporation of multiple copies of a gene, the consequences of gene dosage using the natural promoter can also be pursued. It will also be possible to isolate and investigate the corresponding homologue from animal cells or to even exchange homologues between the two types of cells. The ability to rapidly isolate the entire gene will facilitate identification of noncoding sequence mutations. Because of the ability to manipulate sequences in yeast by recombinational modification, the consequences of specific changes can be investigated in genes that are returned to human cells.
Prior to the present demonstration of direct isolation of a single gene from total DNA, all other methods were laborious because they relied on random isolation of fragmented chromosomal DNA followed by identification of relevant material. Even if a yeast or bacterial artificial chromosome contains an entire gene, the specific recovery of only the gene is an arduous process. Often a gene is available as a set of fragments that forms a contig which must be pieced together, as in the case of BRCA2 (8, 9). The likelihood of isolation of an entire gene by TAR cloning is greatly increased because of the opportunity to rapidly isolate many putative copies of the gene, and it is likely that genes up to several hundred kilobases can be isolated by TAR cloning (5, 6). There are many other utilities of TAR cloning. We have shown (6) that large circular YACs obtained by TAR cloning are stable and can be readily separated from yeast chromosomal DNA. Moreover, circular YACs can be efficiently retrofitted to BACs with a mammalian-selectable marker to enable transfer of YAC/BACs to human cells either directly or after being transformed into E. coli. TAR cloning also provides the opportunity to isolate families of genes, as demonstrated for the human rDNA gene family (16). Families with diverged DNAs could also be isolated because double-strand break recombinational repair can occur between highly diverged DNAs (17–19). Finally, TAR cloning can be used to fill gaps that are present in current genome maps.
Because the entire isolation procedure of chromosomal genes and regions followed by PCR characterization could be accomplished in approximately 2 weeks, the TAR cloning approach can be readily applied to studies of chromosome alterations and human genetic disease. Thus, the TAR cloning approach along with PCR methods will provide powerful tools for genomic analysis and clinical diagnosis.
Acknowledgments
We gratefully acknowledge the advice and critical discussions with Roger Wiseman during many of these experiments and during the development of the manuscript and Jonathan Lancaster for providing the BRCA2 exon 11 PCR primers. The excellent technical assistance of Joan Graves was instrumental in many of the experiments. We appreciate the critical comments of the manuscript by Tom Kunkel. We acknowledge the support of the National Center for Human Genome Research during the early stages of development of the TAR cloning concept, which was initially applied to the general isolation of human genomic DNA and laid the groundwork for the present experiments.
ABBREVIATIONS
- TAR
transformation-associated recombination
- ARS
autonomously replicating sequences
- TAFE
transverse alternating field electrophoresis
- BAC and YAC
bacterial and yeast artificial chromosome, respectively
References
- 1.Claus E B, Risch N, Thompson W D. Am J Hum Genet. 1991;48:232–242. [PMC free article] [PubMed] [Google Scholar]
- 2.Newman B, Austin M A, Lee M, King M-C. Proc Natl Acad Sci USA. 1988;85:3044–3048. doi: 10.1073/pnas.85.9.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wooster R, Neuhausen S L, Mangion J, Quirk Y, Ford D, et al. Science. 1994;265:2088–2090. doi: 10.1126/science.8091231. [DOI] [PubMed] [Google Scholar]
- 4.Friend S. Nat Genet. 1996;13:16–17. doi: 10.1038/ng0596-16. [DOI] [PubMed] [Google Scholar]
- 5.Larionov V, Kouprina N, Graves J, Chen X-N, Korenberg J R, Resnick M A. Proc Natl Acad Sci USA. 1996;93:491–496. doi: 10.1073/pnas.93.1.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larionov V, Kouprina N, Graves J, Resnick M A. Proc Natl Acad Sci USA. 1996;93:13925–13930. doi: 10.1073/pnas.93.24.13925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stinchomb D T, Thomas M, Kelly I, Selker E, Davis R W. Proc Natl Acad Sci USA. 1980;77:4559–4563. doi: 10.1073/pnas.77.8.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tavtigian S V, Simard J, Rommens J, Couch F, Shattuck-Eidens D, et al. Nat Genet. 1996;12:333–337. doi: 10.1038/ng0396-333. [DOI] [PubMed] [Google Scholar]
- 9.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, et al. Nature (London) 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 10.Larionov V, Kouprina N, Nikolaishvili N, Resnick M A. Nucleic Acids Res. 1994;22:4154–4162. doi: 10.1093/nar/22.20.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lancaster J M, Wooster R, Mangion J, Phelan C M, Cochran C, et al. Nat Genet. 1996;13:238–240. doi: 10.1038/ng0696-238. [DOI] [PubMed] [Google Scholar]
- 12.Game J C, Sitney K C, Cook V E, Mortimer R K. Genetics. 1989;123:695–713. doi: 10.1093/genetics/123.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bradshaw M S, Bollekens J A, Ruddle F H. Nucleic Acids Res. 1995;23:4850–4856. doi: 10.1093/nar/23.23.4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ketner G, Spencer F, Tugendreich S, Connelly C, Hieter P. Proc Natl Acad Sci USA. 1994;91:6186–6190. doi: 10.1073/pnas.91.13.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teng D H, Bogden R, Mitchell J, Baumgard M, Bell R, Berry S, Davis T, Ha P C, Kehrer R, Jammulapati S, Chen Q, Offit K, Skolnick M H, Tavtigian S V, Jhanwar S, Swedlund B, Wong A K, Kamb A. Nat Genet. 1996;13:241–244. doi: 10.1038/ng0696-241. [DOI] [PubMed] [Google Scholar]
- 16.Kouprina, N., Graves, G., Resnick, M. A. & Larionov, V. (1997) Gene, in press. [DOI] [PubMed]
- 17.Mezard C, Pompon D, Nicolas A. Cell. 1992;70:659–670. doi: 10.1016/0092-8674(92)90434-e. [DOI] [PubMed] [Google Scholar]
- 18.Larionov V, Kouprina N, Eldarov M, Perkins E, Porter G, Resnick M A. Yeast. 1994;10:93–104. doi: 10.1002/yea.320100109. [DOI] [PubMed] [Google Scholar]
- 19.Priebe S D, Westmoreland J, Nilsson-Tillgren T, Resnick M A. Mol Cell Biol. 1994;14:4802–4814. doi: 10.1128/mcb.14.7.4802. [DOI] [PMC free article] [PubMed] [Google Scholar]


