Abstract
Glycosylphosphatidylinositol (GPI)-anchored proteins are widely distributed on plasma membranes of eukaryotes. More than 50 GPI-anchored proteins have been shown to be spatiotemporally expressed in mice with a deficiency of GPI-anchor biosynthesis that causes embryonic lethality. Here, we examine the functional roles of GPI-anchored proteins in mouse skin using the Cre-loxP recombination system. We disrupted the Pig-a gene, an X-linked gene essential for GPI-anchor biosynthesis, in skin. The Cre-mediated Pig-a disruption occurred in skin at almost 100% efficiency in male mice bearing two identically orientated loxP sites within the Pig-a gene. Expression of GPI-anchored proteins was completely absent in the skin of these mice. The skin of such mutants looked wrinkled and more scaly than that of wild-type mice. Furthermore, histological examination of mutant mice showed that the epidermal horny layer was tightly packed and thickened. Electron microscopy showed that the intercellular space was narrow and there were many small vesicles embedded in the intercellular space that were not observed in equivalent wild-type mouse skin preparations. Mutant mice died within a few days after birth, suggesting that Pig-a function is essential for proper skin differentiation and maintenance.
The core of the glycosylphosphatidylinositol (GPI) anchor consisting of phosphatidylinositol, glucosamine, three mannoses, and ethanolaminephosphate is synthesized in the endoplasmic reticulum and transferred to the C terminus of precursor proteins to form GPI-anchored proteins (1–3). All GPI-anchored proteins share such a common, core GPI anchor. A lack of GPI-anchor biosynthesis, therefore, would cause defective surface expression of many different GPI-anchored proteins and, if caused by a germ line mutation, it would result in lethality at an early development of mouse embryos (4). Paroxysmal nocturnal hemoglobinuria (PNH), an acquired human hematologic disorder, is characterized by deficient expression of GPI-anchored proteins in hematopoietic, but not nonhematopoietic, cells, suggesting that a somatic mutation possibly occurs in a hematopoietic stem cell (5, 6). The gene responsible for PNH is the X-linked gene PIG-A (phosphatidylinositolglycan class A; refs. 7 and 8). Due to X chromosomal inactivation, there is only one functional PIG-A allele in somatic cells; therefore, one somatic mutation in PIG-A would cause GPI-anchor deficiency (8). In contrast, all other genes involved in GPI-anchor biosynthesis have been mapped to autosomes, and, to date, there are no patients with PNH whose GPI-anchor deficiency is caused by mutation in a non-PIG-A gene (9, 10). No inherited human diseases caused by a GPI-anchor deficiency have been reported, probably because a deficiency of all GPI-anchored proteins would cause early embryonic lethality as seen in the mouse.
To circumvent the early embryonic lethality as well as allow the analysis of later functional roles of GPI-anchored proteins during development and maintenance of individual tissue, we utilized the Cre-loxP recombination system for the introduction of a spatiotemporal gene ablation. The system generally requires cross-mating of two lines of genetically manipulated mice. One line of mice carries alleles with a gene of interest flanked by two identically orientated loxP sites. The other line contains a Cre transgene in which the expression of Cre is controlled by a defined promoter. Recombination between the two loxP sites in the mated mice results in the catalysis of a deletion of the region flanked by the loxP sites; this is dependent on Cre transgene expression.
In this study we analyzed functional roles of GPI-anchored proteins in skin development because several GPI-anchored proteins are known to be expressed in skin. To achieve a skin-specific Pig-a (a mouse homologue of PIG-A) disruption, two mouse lines were generated. In one, the Pig-aflox line, loxP sites were inserted 5′ and 3′ of exon 6 of the Pig-a gene. The second line, the keratin 5-Cre (K5Cre) transgenic mice, 14 kb of the keratin 5 promoter was placed upstream of the Cre gene. Because the keratin 5 promoter directs gene expression in basal cells of the skin, Pig-a disruption may occur both in basal cells and progeny of basal cells in offspring between matings of Pig-aflox and K5Cre. The basal cell is thought to be a stem cell in skin; therefore, all skin cells would be expected to be GPI-anchor negative if the Cre-loxP system is working efficiently.
MATERIALS AND METHODS
Construction of Targeting Vectors.
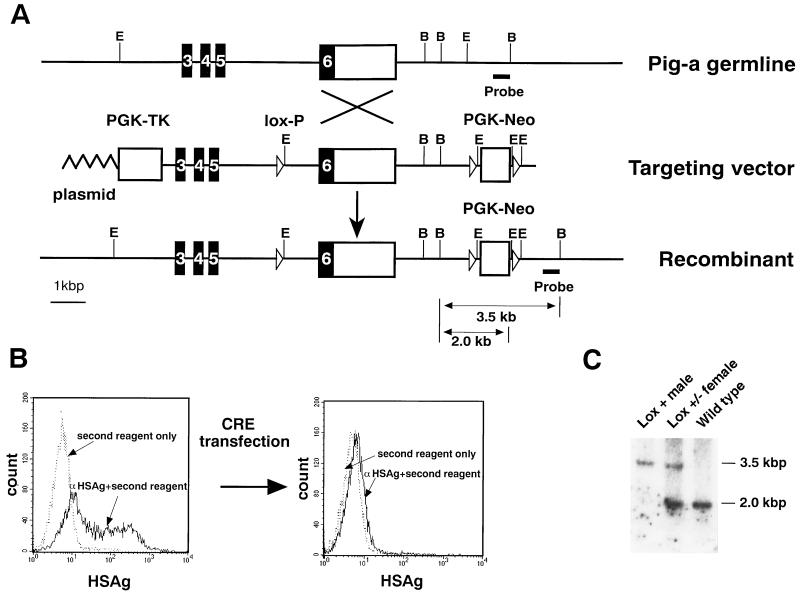
To generate the Pig-a targeting construct, an SalI and blunted SpeI fragment from the mouse genomic Pig-a locus derived from phage clone K1 (11) was inserted into the SalI–SmaI sites of pL2neo (12) after the deletion of the PstI fragment (pPigK1). A blunted XbaI–SalI fragment containing the neomycin resistance gene from pL2neo and an 800-bp PCR fragment derived from downstream of the 3′ EcoRI site (Fig. 1A) were inserted at the EcoRV and SmaI sites of pBluescript II, respectively (pPigneo). An NotI and blunted SalI fragment from pPigneo was inserted into the NotI and blunted EcoRI site of the plasmid containing exon 6 of the genomic Pig-a locus from phage clone K2 (pPigneoK2). A BamHI–XhoI fragment of pPigK1 and SalI–NotI fragment of pPigneoK2 were cloned into the NotI–BamHI site of pBluescript II by triple ligation. PGK-TK from pPNT was then inserted upstream of exon 3 of the genomic Pig-a locus (Fig. 1A).
Figure 1.
Targeted insertion of loxP sites into the Pig-a gene. (A) Part of the wild-type Pig-a locus showing the positions of exons 3–6, the targeting construct, and Pig-a allele containing the introduced loxP sites are shown. Solid and open boxes indicate the coding and noncoding exons, respectively. Restriction sites for BamHI (B) and EcoRI (E) are indicated. (B) Cre-mediated disappearance of GPI-anchored proteins on the surface of ES cells containing the loxP sites in the Pig-a gene. The plasmid pMC-Cre (13) with hygr was transfected into clone 58. The surface expression of heat-stable antigens (HSAg) was examined before and after selection with 150 μg/ml of hygromycin B for 7 days. Five of the 12 clones lost surface GPI-anchored proteins after selection. A representative clone that lost GPI-anchored proteins is shown. (C) Southern hybridization of the DNA from the germ-line-transmitted and control mice. Genomic DNA was obtained from tails and digested with BamHI. The 3.5- and 2.0-kb fragments represent targeted and endogenous alleles, respectively.
Generation and Identification of Targeted Embryonic Stem (ES) Clones.
R1 ES cells (1 × 107) (kindly provided by A. Nagy, Mount Sinai Hospital, Toronto) in 0.8 ml of plain DMEM were transfected with 25 μg of NotI-linearized targeting vector using a Bio-Rad Gene Pulser (set at 210 V and 500 μF). Positive selection with 150 μg/ml of G418 and negative selection with 2 μM of gancyclovir were started from the following day and fourth day after the electroporation, respectively. One week after the electroporation, colonies were picked and analyzed by PCR to detect homologous recombinant clones. PCR was performed with 35 cycles of a reaction consisting of 1 min of denaturation at 93°C, 1 min of annealing at 55°C, and 2 min of elongation at 72°C using two primers, 5′-GGATTTATTAAAATGCCTTACAGG in the downstream region of exon 6 of the Pig-a gene, and 5′-GGGTTATTGAATATGATCGGAATT in the upstream of the Neo resistance gene. PCR was carried out in the following mixture with a final volume of 50 μl: 30 mM tricine, pH 8.4/2 mM MgCl2/5 mM 2-mercaptoethanol/0.01% gelatin/0.01% Triton X-100/2.5 mM of each dNTP/1 μM of each primer/2 units of Taq DNA polymerase.
K5 Promoter Activity and Generation of K5 Cre Transgenic Mice.
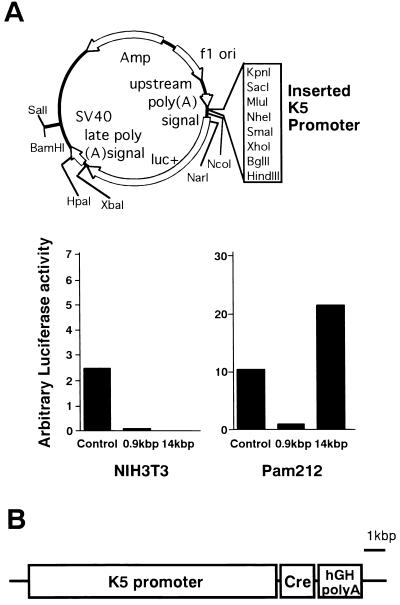
A 14-kb fragment of the K5 promoter was cloned from a human genomic library in phage λFix II. A 0.9-kb blunted SpeI–BamHI fragment was inserted into the SmaI site of pGL3-Basic Vector (Promega), and a 14-kb blunted SpeI and KpnI fragment of K5 promoter was inserted into SmaI and KpnI sites of the pGL3-Basic Vector (Fig. 2A). Fifteen micrograms of these plasmids was electroporated into a mouse keratinocyte cell line (PAM 212) (14) and fibroblasts (NIH 3T3) in DMEM with 10% fetal calf serum using a Bio-Rad gene pulser (950 μF and 250 V). After 2 days luciferase activity was measured using a luminescence reader (Aloka, Mitaka, Tokyo). Promoter activities were compared with pGL3 Control Vector (Promega). Luciferase activities are indicated as arbitrary units. A Cre cDNA with the nuclear localizing signal was cloned into pBluescript (pBSCre). An NotI and blunted BamHI fragment of pUC1017CreK (a gift from J. Marth, University of California at San Diego, La Jolla) that contained human growth hormone poly(A) was inserted into the NotI and blunted HindIII sites of pBSCre (pBSCreA). An SpeI–SalI fragment of the K5 promoter was subcloned into SpeI–SalI sites of pBluescript II (pBSK5). An NotI and blunted XhoI fragment of pBSCreA was inserted into the NotI and blunted SpeI sites of pBSK5. This plasmid was linealized with KpnI–NotI and injected into fertilized eggs after purification by DE81 paper. Fertilized eggs collected from the oviducts of B6C3F1 females mated with B6C3F1 males were injected according to standard procedures, and two founder mice were born. Founder mice and their littermates were screened by PCR. PCR was performed with genomic DNA from mouse tail templates under the following conditions: a 5′-specific primer (primer Kr1) of K5 promoter (5′-ATGCCAATGCCCCCTCAGTTCCT), and a 3′-specific primer (primer Kr2) of K5 promoter (5′-TGCCCCTTTTTATCCCTTCCAGA) with 35 cycles of a reaction consisting of 1 min of denaturation at 93°C, 1 min of annealing at 57°C, and 2 min of elongation at 72°C.
Figure 2.
Creation of the K5Cre transgenic lines. (A) K5 promoter activity in cultured cell lines. The construct in which the luciferase gene was placed under the control of the K5 promoter (Upper) was transfected into a keratinocyte cell line (PAM 212) and a fibroblast line (NIH 3T3). The control vector in which the luciferase gene was driven by an simian virus 40 early promoter and its enhancer was also transfected into these cell lines. The luciferase activities were measured and are indicated as arbitrary units (Lower). (B) Structure of the K5Cre transgene. A 14-kb fragment of human K5 promoter used to control the expression of Cre transgene and the nuclear localizing signal was added at 5′ of Cre gene.
PCR Analysis of Tissue-Specific Disruption of the Pig-a Gene.
PCR was performed with genomic DNA (200 ng) from various tissues as templates under the following conditions: a 5′-specific primer (primer 1) upstream to 5′ site in the intron 5 (5′-ACCTCCAAAGACTGAGCTGTTG), a 3′-specific primer (primer 2) just downstream of 5′ loxP site in intron 5 (5′-CCTGCCTTAGTCTTCCCAGTAC), and a 3′-specific primer downstream of Pig-a gene (5′-TGTGGGTTTCAGTTCATTTCAGA) with 28 cycles of a reaction consisting of 1 min of denaturation at 93°C, 1 min of annealing at 65°C, and 2 min of elongation at 72°C. To separate keratinocytes from fibroblasts, skin from newborn mice was incubated in 0.25% trypsin in PBS at 4°C overnight.
Flow Cytometric Analysis of Keratinocytes.
Skin was removed from newborn mice and incubated in 0.25% trypsin in PBS at 4°C overnight to separate keratinocytes from fibroblasts. Flow cytometric analysis was performed by using anti-HSAg as described previously (4).
Histological and Electron Microscopic Analysis.
Skin was fixed in 10% neutral formaldehyde solution and embedded in paraffin. Sections were cut at 2 μm and stained with hematoxylin-eosin. For electron microscopy, mouse skin was minced into 1-mm cubes and fixed immediately in 5.0% glutaraldehyde/0.2 M sodium acetate/lead nitrate, and examined under a H-7100 electron microscope.
RESULTS AND DISCUSSION
Generation of ES Cells with a Defined Deletion at the Pig-a Locus and Generation of Chimeric Mice.
To avoid early embryonic lethality and determine the functions of GPI-anchored proteins in individual tissues or organs, we utilized the Cre-loxP system (15) for the disruption of the Pig-a gene. The Cre-loxP system is a bipartite system requiring a Cre-expressing mouse line and a target mouse line for Cre in which loxP sites are inserted into or flank a target gene. Our target was the X-linked Pig-a gene, the mouse homologue of human PIG-A (11). To generate a targeting construct, one loxP site was inserted into intron 5 of the gene, and two additional loxP sites flanking the neomycin resistance gene (Neo) were placed downstream of the Pig-a gene (Fig. 1A). Concomitant with expression of the Cre recombinase, exon 6 of Pig-a together with Neo are expected to be deleted. The targeting construct was transfected into R1 ES cells. We screened 296 colonies, and 12 PCR-positive clones were identified after G418 and gancyclovir selection. Ten of these PCR-positive clones did not retain the 5′ loxP site located within intron 5 (Fig. 1A). One of the clones bearing a 5′ loxP site (clone 58) had the correctly altered Pig-a locus as judged by Southern blot analysis (data not shown).
We first tested whether these loxP sites are recognized by the Cre recombinase. This was achieved by transfecting a Cre-encoding plasmid DNA (pMC-HygCre) into clone 58, Pig-afloxES cells. Because Pig-a is on the X chromosome and R1 ES cells are of male origin, expression of GPI-anchored proteins on the Pig-afloxES cell surface should become deficient if the loxP sites are recognized and a recombination event is mediated by the Cre recombinase. The insertion of loxP sites and the Neo gene per se did not affect the expression of GPI-anchored proteins examined in the Pig-afloxES cells prior to the introduction of the Cre protein (data not shown). As shown in Fig. 1B, the surface expression of heat-stable antigen, a GPI-anchored protein, on Pig-afloxES cells disappeared after pMC-HygCre transfection and selection in hygromycin. This suggests that the loxP sites were indeed recognized in the Pig-afloxES cells. Chimeric mice were generated from Pig-afloxES cells, and germ-line-transmitted mice were then obtained (Pig-aflox). Genomic DNAs from these mice were analyzed by Southern blot analysis (Fig. 1C). Because the Pig-a gene is localized on X chromosome, a male mouse bearing a loxP site had only targeted Pig-a locus (Fig. 1C).
Generation of Skin-Specific Cre Transgenic Mice.
In this study, the Cre protein was expressed in the basal cells of the skin, and, consequently, the Pig-a gene was disrupted at this site. Because the basal cells differentiate to the spinous cells, the disruption of the Pig-a gene would cause a complete deficiency of GPI-anchored proteins in the epidermis. Such a skin-specific disruption was accomplished by constructing Cre transgenes driven by the keratin 5 (K5) promoter. The function and specificity of the promoter was confirmed by transfection of the Cre construct into a mouse keratinocyte cell line (PAM 212) (14) and into fibroblasts (NIH 3T3). The 14-kb fragment conferred high luciferase activity in PAM 212, but not in NIH 3T3 cells (Fig. 2A). Cre transgenic mice were then produced using this 14-kb K5 promoter (K5Cre; Fig. 2B).
Generation of Skin-Specific Pig-a Knockout Mice.
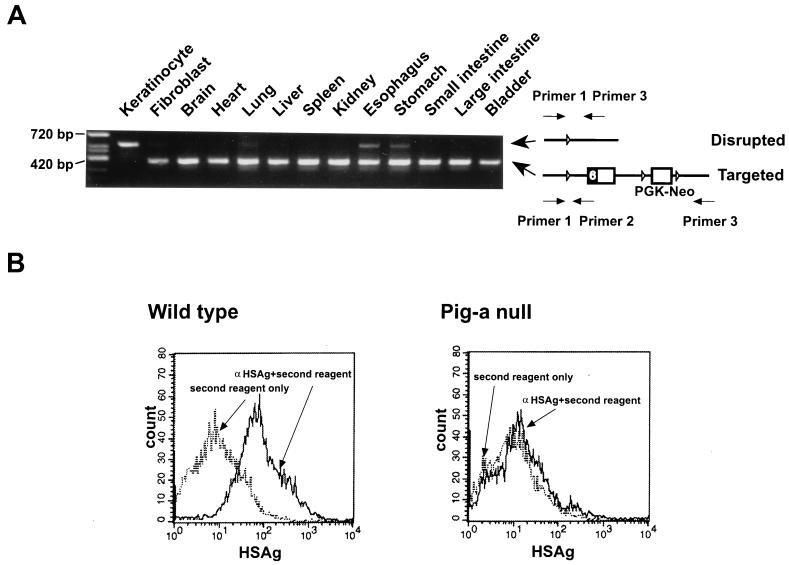
Pig-aflox female (both Pig-a alleles contain loxP) and K5Cre male mice were mated to analyze the function of GPI-anchored proteins in the skin. First, disruption of the Pig-a gene in various tissues of the Cre-positive male offspring (K5Cre:Pig-aflox) was assessed by specific PCR, as shown in Fig. 3A, using three primers. The 5′ primer (primer 1) binds to genomic DNA regardless of the disruption of Pig-a gene (Fig. 3A). Primer 2 binds to the targeted, but not to the disrupted, configuration. Although primer 3 binds to both, it works only on the disrupted configuration, because the distance between primers 1 and 3 is relatively large (≈5 kb) in the targeted configuration. The Pig-a disruption occurred exclusively in epidermis at almost 100% efficiency because the band indicative of the targeted configuration completely disappeared, and that of disruptant emerged (Fig. 3A). Some disruptions were also observed in the esophagus and stomach, suggesting that the K5 promoter was also active in these tissues, which is consistent with previous reports that the K5 gene is expressed not only in epidermis, but also in the esophagus and stomach (16, 17). As expected, the expression of GPI-anchored proteins was completely deficient in K5Cre:Pig-aflox male epidermis (Fig. 3B). In contrast, half of the epidermal cells from K5Cre:Pig-aflox/W female mice (only one Pig-a allele contained loxP) expressed GPI-anchored proteins due to the random inactivation of X chromosomes (data not shown). Although 27 out of 37 K5Cre:Pig-aflox male mice died at 1–3 days after birth, the remaining 10 mice survived. We examined the disruption of Pig-a gene in the epidermis in the surviving mice and found that the expected disruption had not occurred in epidermis (data not shown). This fact suggests that Cre expression in the surviving mice was not high enough to recognize loxP sites and disrupt Pig-a gene.
Figure 3.
Tissue-specific disruption of the Pig-a gene and deficient expression of GPI-anchored proteins in Cre:Pig-aflox male mice. (A) Tissue-specific disruption of the Pig-a gene in Cre:Pig-aflox male mice. To estimate the efficiency of Pig-a disruption, allele-specific PCR was performed using the three primers shown on the right. Size markers are included. (B) Deficient expression of a GPI-anchored protein, heat-stable antigen (HSAg), in keratinocytes. Epidermis from 1-day-old mice was separated from dermis by trypsin, and then individual cells were obtained. The surface expression of HSAg was examined.
Analysis of Skin-Specific Pig-a Knockout Mice.
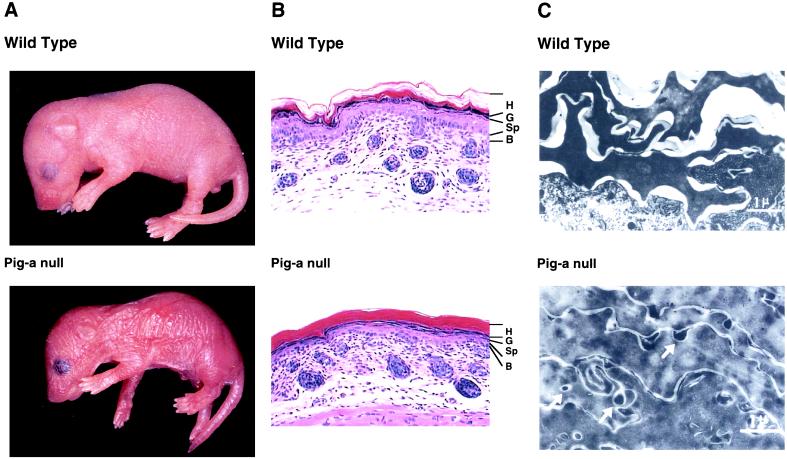
The appearance of the mice was compared with that of wild-type mice in Fig. 4A. The skin of K5Cre:Pig-aflox male mice looked wrinkled and more scaly than that of wild-type counterparts. Most K5Cre:Pig-aflox/W female mice survived, but scaly skin appeared in these mice at about 4 days after birth (data not shown). Histological examination of the K5Cre:Pig-aflox male mice showed that the epidermal horny layer (H) was tightly packed and thickened (Fig. 4B). There was no significant difference in the granular (G), spinous (Sp), and basal (B) layers of the epidermis compared with those of the wild-type mice (Fig. 4B). Immunohistochemical examination with anti-filaggrin and anti-loricrin antibodies demonstrated that the staining profiles of the granular cells did not differ between K5Cre:Pig-aflox male mice and wild-type counterparts (data not shown). The structure of the hair follicles was also normal (Fig. 4B). The epithelial cells from esophagus and stomach of K5Cre:Pig-aflox male mice where the Pig-a gene was partially disrupted were examined. We did not find any changes in these tissues, suggesting that the primary reason for the lethality would be a defect in the skin (data not shown). K5Cre:Pig-aflox/W female mice exhibited only slight acanthosis compared with wild-type mice (data not shown). Fig. 4C shows electron microscopic examination of the horny layer. In wild-type mice it comprises amorphous cornified cells consisting mainly of protein components such as keratin and cystatin and an intercellular space where lipid components such as ceramide and cholesterol are packed and then extracted by organic solvents during sample preparation for electron microscopy. In K5Cre:Pig-aflox male mice, the intercellular space was narrow and there were many small vesicles embedded in an amorphous substance that was not seen in wild-type mice (Fig. 4C). These results suggest that lipid components in lamellar granules either are not secreted into the extracellular space or secreted but inefficiently rearranged in the intercellular space, thereby perturbing normal development in the epidermis of the K5Cre:Pig-aflox male mouse. The lipid rearrangement after the secretion from lamellar granules is important for the epidermal barrier formation (18). A lack of lipid components in the intercellular space may cause a severe water loss through the epidermis and lead to neonatal lethality in mice. It is known that the apical sorting of GPI-anchored proteins in polarized cells is blocked by an inhibitor of ceramide biosynthesis, suggesting that GPI-anchored proteins and ceramide in vesicles are cotransported from the Golgi apparatus to the apical plasma membrane (19). Due to a lack of GPI-anchored proteins in the Cre:Pig-aflox male epidermis, the transport of ceramide, which is the main component of the lamellar granules, to the plasma membrane would be blocked, thereby impairing the release of lipid components to the extracellular space. A similar phenotype with an abnormal horny layer has been described as type I harlequin ichthyosis (20). Because mice carrying a complete disruption of the X-linked Pig-a gene exhibit deficient expression of all GPI-anchored proteins on the cell surface, leading to lethality at an early embryonic stage, the function of GPI-anchored proteins could not be analyzed at later developmental stages in such animals.
Figure 4.
Macro- and microscopic changes of epidermis from wild-type and skin-specific, Pig-a-disrupted mouse (Cre:Pig-aflox male mouse). (A) Macroscopic appearance of the wild-type and the skin-specific, Pig-a-disrupted mouse. (B) Histological examination of the epidermis from the wild-type and the skin-specific, disrupted mouse. (Hematoxylin-eosin staining, ×200.) (C) Histological examination of the horny layers by electron microscopy. The vesicles in the horny layers are indicated by white arrows.
This study demonstrates that a conditional knockout utilizing the Cre-loxP system permitted analysis of the function of GPI-anchored proteins in specific tissues. It was shown that a cell-type-specific gene targeting using the Cre-loxP system could be a potential tool for analyzing the tissue-specific function of a gene of interest, especially when a null mutation of that gene in the germ line causes lethality at an early embryonic stage (15, 21). The present study demonstrates that the Cre-loxP strategy works in vivo for disclosing specific function(s) of a gene of interest. The disruption of the Pig-a gene was almost complete and exclusively confined to skin judged by allele-specific PCRs and the analysis of the expression of GPI-anchored proteins in the epidermis. This tissue-specific knockout of the Pig-a gene has clarified that GPI-anchored proteins are involved in lipid (ceramide) transport into extracellular space of the horny layer of the epidermis, although it is not known whether this finding is a direct or indirect consequence of the Pig-a gene knockout. A line of study as that taken here could facilitate the functional analysis of GPI-anchored proteins in many other tissues when tissue-specific Cre transgenics are available.
Acknowledgments
We thank Drs. H. Kondo, A. Nagy, and K. Hadjantonakis for critical reading of the manuscript and Drs. W. Müller and K. Rajewsky for providing the Cre-loxP constructs and encouraging us during the initial stages of this project. This work was supported by grants from the Ministry of Education, Science, Sports, and Culture of Japan.
ABBREVIATIONS
- GPI
glycosylphosphatidylinositol
- PNH
paroxysmal nocturnal hemoglobinuria
- PIG-A
phosphatidylinositolglycan class A
References
- 1.Stevens V L. Biochem J. 1995;310:361–370. doi: 10.1042/bj3100361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Udenfriend S, Kodukula K. Annu Rev Biochem. 1995;64:563–591. doi: 10.1146/annurev.bi.64.070195.003023. [DOI] [PubMed] [Google Scholar]
- 3.Takeda J, Kinoshita T. Trends Biochem Sci. 1995;20:367–371. doi: 10.1016/s0968-0004(00)89078-7. [DOI] [PubMed] [Google Scholar]
- 4.Kawagoe K, Kitamura D, Okabe M, Taniuchi I, Ikawa M, Watanabe T, Kinoshita T, Takeda J. Blood. 1996;87:3600–3606. [PubMed] [Google Scholar]
- 5.Kinoshita T, Inoue N, Takeda J. Adv Immunol. 1995;60:57–103. doi: 10.1016/s0065-2776(08)60584-2. [DOI] [PubMed] [Google Scholar]
- 6.Rosse W F, Ware R E. Blood. 1995;86:3277–3286. [PubMed] [Google Scholar]
- 7.Miyata T, Takeda J, Iida Y, Yamada N, Inoue N, Takahashi M, Maeda K, Kitani T, Kinoshita T. Science. 1993;259:1318–1320. doi: 10.1126/science.7680492. [DOI] [PubMed] [Google Scholar]
- 8.Takeda J, Miyata T, Kawagoe K, Iida Y, Endo Y, Fujita T, Takahashi M, Kitani T, Kinoshita T. Cell. 1993;73:703–711. doi: 10.1016/0092-8674(93)90250-t. [DOI] [PubMed] [Google Scholar]
- 9.Ware R E, Howard T A, Kamitani T, Chang H M, Yeh E T H, Seldin M F. Blood. 1994;83:3753–3757. [PubMed] [Google Scholar]
- 10.Kinoshita T, Inoue N, Takeda J. Annu Rev Med. 1996;47:1–10. doi: 10.1146/annurev.med.47.1.1. [DOI] [PubMed] [Google Scholar]
- 11.Kawagoe K, Takeda J, Endo Y, Kinoshita T. Genomics. 1994;23:566–574. doi: 10.1006/geno.1994.1544. [DOI] [PubMed] [Google Scholar]
- 12.DiSanto J P, Müller W, Guy-grand D, Fischer A, Rajewsky K. Proc Natl Acad Sci USA. 1995;92:377–381. doi: 10.1073/pnas.92.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu H, Zou Y, Rajewsky K. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- 14.Yuspa S H, Hawley-Nelson P, Koehler B, Stanley J R. Cancer Res. 1980;40:4694–4703. [PubMed] [Google Scholar]
- 15.Gu H, Marth J D, Orban P C, Mossmann H, Rajewsky K. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 16.Byrne C, Fuchs E. Mol Cell Biol. 1993;13:3176–3190. doi: 10.1128/mcb.13.6.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramirez A, Bravo A, Jorcano J L, Vidal M. Differentiation. 1994;58:53–64. doi: 10.1046/j.1432-0436.1994.5810053.x. [DOI] [PubMed] [Google Scholar]
- 18.Ebling F J G. In: Textbook of Dermatology. Champion R H, Burton J L, Ebling F J G, editors. Oxford: Blackwell Scientific; 1992. pp. 125–155. [Google Scholar]
- 19.Mays R W, Siemers K A, Fritz B A, Lowe A W, van Meer G, Nelson W J. J Cell Biol. 1995;130:1105–1115. doi: 10.1083/jcb.130.5.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dale B A, Holbrook K A, Fleckman P, Kimball J R, Brumbaugh S, Sybert V P. J Invest Dermatol. 1990;94:6–18. doi: 10.1111/1523-1747.ep12873301. [DOI] [PubMed] [Google Scholar]
- 21.Tsien J Z, Chen D F, Gerber D, Tom C, Mercer E H, Anderson D J, Mayford M, Kandel E R, Tonegawa S. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]