Abstract
In a cell line (NB4) derived from a patient with acute promyelocytic leukemia, all-trans-retinoic acid (ATRA) and interferon (IFN) induce the expression of a novel gene we call RIG-G (for retinoic acid-induced gene G). This gene codes for a 58-kDa protein containing 490 amino acids with several potential sites for post-translational modification. In untreated NB4 cells, the expression of RIG-G is undetectable. ATRA treatment induces the transcriptional expression of RIG-G relatively late (12–24 hr) in a protein synthesis-dependent manner, whereas IFN-α induces its expression early (30 min to 3 hr). Database search has revealed a high-level homology between RIG-G and several IFN-stimulated genes in human (ISG54K, ISG56K, and IFN-inducible and retinoic acid-inducible 58K gene) and some other species, defining a well conserved gene family. The gene is composed of two exons and has been mapped by fluorescence in situ hybridization to chromosome 10q24, where two other human IFN-stimulated gene members are localized. A synergistic induction of RIG-G expression in NB4 cells by combined treatment with ATRA and IFNs suggests that a collaboration exists between their respective signaling pathways.
Acute promyelocytic leukemia (APL) is the first example of human cancer differentiation therapy (1) and is characterized by a specific chromosome translocation t(15;17) that fuses the retinoic acid receptor (RAR) α gene on chromosome 17q21 with the promyelocytic leukemia (PML) gene on chromosome 15q22 (2). The chimeric PML-RARα receptor retains all the important functional domains of both genes. It was previously considered that PML-RARα inhibits all-trans-retinoic acid (ATRA)-induced differentiation and contributes to leukemogenesis in a dominant-negative fashion and that ATRA treatment converts PML-RARα from an antagonist to an agonist of differentiation (3). Recent studies indicate that ATRA can increase the degradation of PML-RARα and allows APL cells to differentiate by relieving the differentiation block caused by PML-RARα (4, 5). The molecular mechanism(s) that might lead to the functional conversion of PML-RARα or to its degradation by ATRA treatment leading to terminal differentiation are not yet clarified.
During ATRA-induced differentiation of APL cells, a vast array of genes may be regulated (6). By using differential display–PCR (DD-PCR) technique, a gene designated RIG-G has been shown to be expressed in NB4 cells induced to differentiate by ATRA. Because RIG-G is also inducible by interferon (IFN) and bears strong homology to a family of the IFN-stimulated gene (ISG), a possible crosstalk between retinoid and IFN signaling pathways has been speculated and further examined.
MATERIALS AND METHODS
Cell Culture and Treatment.
APL cell line NB4 and breast cancer cell lines T47D and MDA-MB-231 were cultured in RPMI medium 1640 supplemented with 10% calf serum. ATRA (10−6 to 10−8 M) was added to cell culture with or without human recombinant IFN-α or IFN-γ (500 units/ml and 2,000 units/ml, respectively). In some experiments, cells were treated only with IFNs.
DD-PCR.
DD-PCR was performed as previously described (6) with the following modification. The total RNA of NB4 cells was pretreated with DNase I (RNase-free) to eliminate DNA contamination. DD-PCR products were separated by sequencing gel and visualized by autoradiography. The reproducible bands were excised and eluted from the gel. After reamplification, the bands were cloned into pGEM-T vector (Promega).
cDNA and Genomic DNA Cloning.
The cDNA library was constructed from the mRNAs of NB4 cells treated with ATRA for 48 hr. The cDNA fragment from DD-PCR was used to screen the library according to the standard protocol (Stratagene). One of the cDNA clones isolated from cDNA library was prepared as a probe to screen the human genomic DNA library constructed into EMBL3 (CLONTECH).
DNA Sequencing and Sequence Analysis.
DNA sequencing was carried out using T7 Sequencing Kit (Pharmacia) or performed on 373 DNA sequencer using cycle sequencing kit (Applied Biosystems). The complete cDNA sequence was constructed by sequencing the overlapping clones and primer extension. The genomic clones isolated were characterized by restriction map and Southern blot analysis and were subcloned into pBluescript and partially sequenced. The cDNA sequence was analyzed by using stride 1.2 software. The sequence of cDNA and the predicted protein sequence were searched against GenBank and Swissprot databases for homology using the GCG package (Genetics Computer Group, Madison, WI).
Northern Blot Analysis.
Total RNA (20 μg) was size-fractionated on 1.2% agarose/5% formamide and transferred to nylon membrane by capillary method. The Northern blot I containing mRNAs from eight different tissues (heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas) was from CLONTECH. Hybridization was performed by using 1.5-kb cDNA probe under the conditions reported previously (6).
In Vitro Transcription and Translation.
The cDNA containing the whole ORF was purified and used as template for in vitro transcription and translation, by using TNT Coupled Reticulocytic Lysate System (Promega) using [35S]methionine according to manufacturer’s recommendation. The proteins were separated on PAGE and visualized by autoradiography.
Expression Plasmid Construction.
An epitope-tagged expression vector was constructed by insertion of a duplex oligonucleotide sequence: 5′-TCGAGGCCACCATGGACTACAAGGACGACGATGACAAGGTAC-3′ 3′-CCGGTGGTACCTGATGTTCCTGCTGCTACTGTTC-5′ into pCI (Promega) with XhoI and KpnI sites (7). A primer pair was designed to amplify the entire RIG-G coding sequence. The resultant PCR fragment has a KpnI site on the 5′ end and is in frame with the Flag epitope sequence. The fragment then was inserted into the constructed epitope-tagged pCI vector using KpnI and SmaI sites. The resulting fusion gene encodes the RIG-G protein, with N-terminal Flag epitope.
Transfection and Immunofluorescent Cell Staining.
Chinese hamster ovary cells were used for transient transfection assay to determine the subcellular location of RIG-G protein. The transfection assay was performed using a LipofectAmine Transfection Kit (GIBCO/BRL) according to the manufacturer’s instructions. The cells transfected with RIG-G expression vector were fixed and incubated with anti-Flag mAb M2 (IBI), followed by incubation with fluorescein isothiocyanate-conjugated anti-mouse IgG.
Fluorescence in Situ Hybridization.
Two phage clones, about 15 kb in length, were isolated from the EMBL3 genomic library and used as probes. The phage DNAs were biotinylated with biotin-16-dUTP using nick translation system (Boehringer Mannheim) and probed on normal human lymphocyte chromosomes in metaphase (8). The hybridization signal and PI (Sigma)-banded chromosomes were visualized with a Nikon microscope and merged using Cytovision image analysis software (Applied Imaging, Sunderland, U.K.).
Yeast Artificial Chromosome (YAC) Library Screening.
To facilitate the mapping of RIG-G with regard to other members of the ISG family, the Centre d’Etude du Polymorphisme Humain (CEPH) MegaYAC library was screened with PCR using specific primers for RIG-G. Two YAC clones, namely 638E08 and 944C01, were obtained and analyzed using Southern analysis and PCR for human ISG-54K and ISG-56K genes.
RESULTS
Isolation and Characterization of RIG-G cDNA.
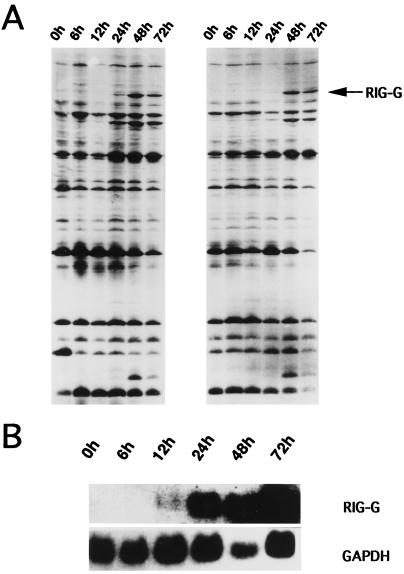
DD-PCR was carried out to identify ATRA-induced genes. In the 30 primer pair combinations, one pair of primers, 3′DD6 (TTTTTTTTTTTCC) and 5′DD7 (TACAACGAGG), produced three differentially displayed bands in NB4 cells treated with ATRA (Fig. 1A). The resulting pattern was reproducible, and the differentially amplified products were excised and cloned. One clone (600 bp in length) was confirmed to be part of a differentially expressed gene by Northern blot analysis (Fig. 1B). This cloned PCR product was then sequenced. After searching through GenBank with blast, this cDNA fragment proved to be from a novel gene, which was then named RIG-G (for retinoic acid-induced gene G).
Figure 1.
Induced expression of RIG-G by ATRA. (A) Differential display of mRNA from NB4 cells treated with ATRA for 6, 12, 24, 48, and 72 hr. Untreated cells (0h) are used as control. RIG-G is derived from the band indicated by the arrow. (B) Northern blot analysis of RIG-G expression in NB4 cells treated with ATRA (10−6 M) for 6, 12, 24, 48, and 72 hr. Hybridization with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA is a control for the amount of RNA loaded.
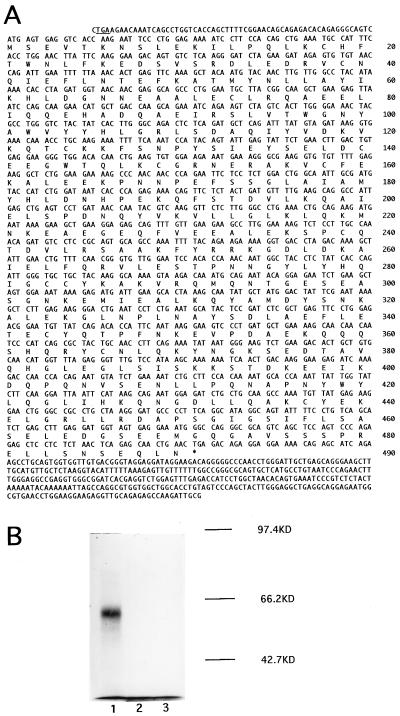
To get the full-length cDNA, a cDNA library of NB4 cells treated with ATRA for 48 hr was constructed with random primers and screened with this novel cDNA fragment obtained from DD-PCR. Eighty positive clones were identified, among which eight cDNA clones were isolated. These clones collectively define a 1,919-bp cDNA sequence (Fig. 2A), which contains a 1,473-bp ORF encoding 490 amino acid residues. A stop codon is found in frame upstream of the first ATG codon (Fig. 2A). The amino acid sequence deduced from the 1,919-bp RIG-G cDNA is shown in Fig. 2A. The in vitro transcription and translation of RIG-G gene revealed a protein product of 58 kD in good agreement with the predicted amino acid sequence of cDNA (Fig. 2B).
Figure 2.
Characterization of RIG-G cDNA. (A) RIG-G cDNA nucleotide sequence and deduced amino acid sequence. The amino acid sequence is listed below the nucleotide sequence and numbered on the right. The termination codon in frame upstream of the start codon is underlined. (B) In vitro transcription and translation of RIG-G cDNA. The TNT Coupled Reticulocytic Lysate System was supplemented with 1.5-kb RIG-G cDNA using T3 (lane 1) or T7 (lane 2) RNA polymerase, or without template (lane 3). The size markers are listed on the right.
RIG-G Is Highly Homologous to Some Members of ISGs.
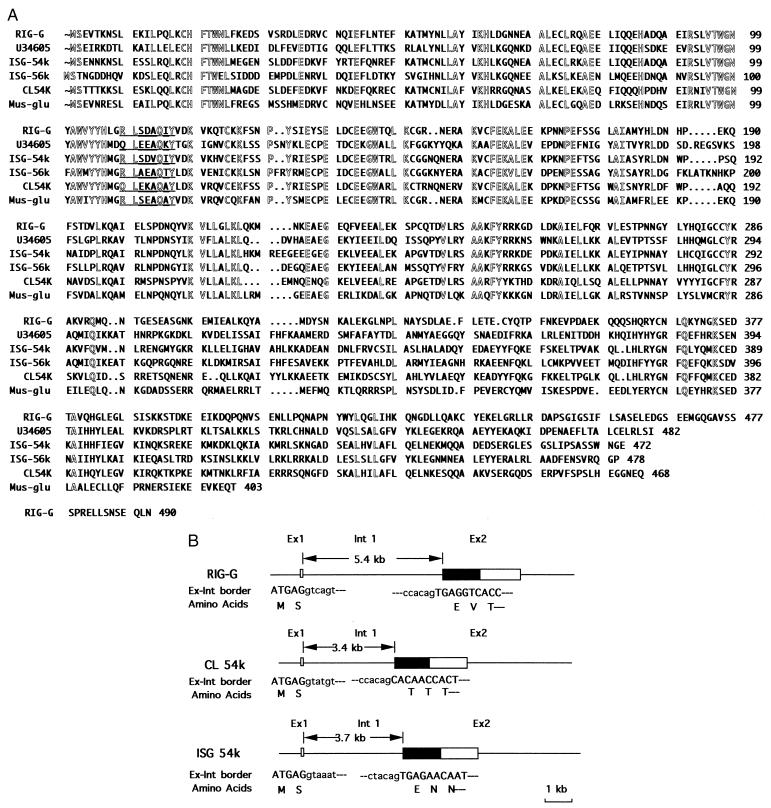
Computer-assisted homology comparison revealed that the RIG-G shares high homology with some members of ISGs. Deduced amino acid sequences of those proteins are of high similarity. For example, the homology of deduced RIG-G protein sequence with the protein sequences deduced from human ISG 54K, human ISG 56K, human IFN-inducible and retinoic acid-inducible 58K (U34605), C.L ISG 56K and murine IRG2 are 57.3% (473 amino acids), 44.3% (470 amino acids), 44.0% (461 amino acids), 50.5% (424 amino acids), and 60.3% (403 amino acids), respectively (Fig. 3A) (9–12). The amino acid sequence at the N terminal is more conserved than that at the C terminal, suggesting that the N-terminal region is more important for function.
Figure 3.
Homology of RIG-G with related ISGs. (A) Comparison of deduced protein sequence of RIG-G with those of related ISG proteins. Amino acid residues conserved in the six ISGs are outlined. A potential tyrosine phosphorylation site is underlined. The numbers on the right represent the amino acid residues of the corresponding protein. (B) The exon-intron organization of RIG-G-related family members. The boxes represent the exons of the genes, and the coding regions of the genes are designated by filled boxes. The length of intron 1 is indicated by arrowheads. The exon-intron border sequences are listed below the exon-intron organization. The nucleotides of exon and intron are shown in capital and lowercase letters, respectively, and the corresponding amino acid residues are listed below the exon sequences.
Several potential sites for post-translational modification are present in the deduced RIG-G protein sequence, such as one potential N-glycosylation site (at residue 405), one glycosaminoglycan attachment site (at residue 451), two potential cAMP- and cGMP-dependent kinase phosphorylation sites (at residues 125 and 391), six casein kinase II phosphorylation sites (at residues 136, 394, 459, 461, 478, and 484), and five protein kinase C phosphorylation sites (at residues 123, 318, 390, 394, and 478). In addition, a potential tyrosine phosphorylation site at residue 109 is conserved among all RIG-G-related family members.
Expression Pattern of RIG-G Gene.
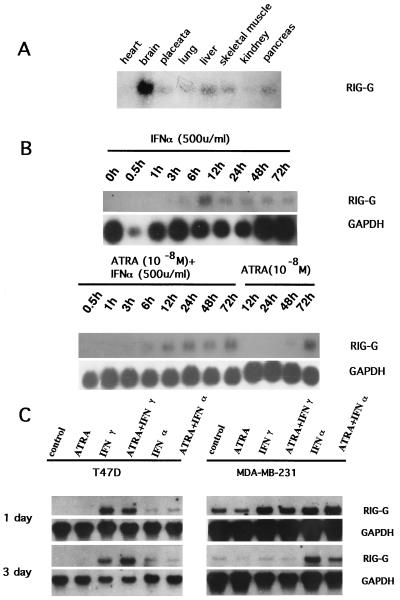
The Northern blot analysis detected an unique transcript of 2.8–2.9 kb in NB4 cells treated with ATRA but not in untreated NB4 cells. The kinetic study showed that RIG-G expression was detectable at 12–24 hr after ATRA (10−6 M) treatment and reached a maximal level at 48–72 hr (Fig. 1B). When the concentration of ATRA was decreased to 10−8 M, the expression of RIG-G could still be induced but at relatively lower level and was detected at a later stage (24–48 hr after ATRA treatment) (Fig. 4B). The induction may require protein synthesis, because it was inhibited by cycloheximide (data not shown), indicating that RIG-G expression is dependent on at least one ATRA-induced protein. IFN-α (500 units/ml) was also capable of inducing RIG-G expression in NB4 cells but with different kinetics. The expression rapidly increased at 30 min to 3 hr and reached its peak at 6 hr followed by slow declines. When NB4 cells were treated with both ATRA and IFN-α, there was a high level expression of RIG-G from 6 to 72 hr (Fig. 4B). IFN-γ alone (2,000 units/ml) exerted no effect on the expression of RIG-G in NB4 cells. Nevertheless, when it was used with ATRA (10−8 M), the expression of RIG-G was earlier and enhanced (data not shown). These data taken as a whole suggest a synergy between ATRA and IFNs in inducing RIG-G expression.
Figure 4.
Pattern of RIG-G expression. (A) The expression of RIG-G mRNA in eight tissues. (B) The time course of RIG-G mRNA induced by IFN-α (500 units/ml), ATRA (10−8 M), or the combination. The time course of NB4 cells treated by ATRA or IFN-α is indicated (Upper). (Lower) The same blot probed with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA. (C) The expression of RIG-G in ATRA-sensitive and ATRA-resistant breast cancer cell lines (T474D and MDA-MB-231, respectively). The reagents used are indicated above. The time courses of the treatment are listed on the left. The hybridization with GAPDH cDNA was used as the internal control of the amount of the RNA loaded.
In contrast to the induced expression in NB4 cells, no obvious expression of RIG-G was observed in HL-60 before and after ATRA treatment. In addition, no RIG-G mRNA was detected in fresh cells from patients with acute myeloid and lymphoid leukemias as well as the U-937 cell line (data not shown). The basal level of expression of RIG-G was low in eight normal human tissues studied (Fig. 4A). Hybridization signals could be detected by Northern blot using poly(A)+ RNA only after prolonged exposure (15 days). Although RIG-G expression could be induced by IFN-γ or IFN-α in two breast cancer cell lines (ATRA proliferation sensitive and resistant breast cancer cell lines T47D and MDA-MB-231), no significant modulation of expression was observed when these cells were treated with ATRA (Fig. 4C).
Subcellular Distribution of RIG-G Protein.
Transfection performed in Chinese hamster ovary cells using Flag-tagged RIG-G expression vector, followed by anti-flag mAb M2 and indirect immunofluorescence staining showed a diffuse distribution pattern in the cytoplasm, with an enhancement in the perinuclear region. There was no obvious nuclear staining (Fig. 5).
Figure 5.

(A) Control Chinese hamster ovary cells transfected with pCI vector. (B) The subcellular localization of RIG-G. Note that the expressed proteins are distributed in the cytoplasm, especially in the perinuclear region.
Genomic Structure and Chromosomal Localization of RIG-G Gene.
A genomic library in EMBL3 was screened with 1.5-kb RIG-G cDNA fragment. Five positive clones were isolated and characterized by restriction map and Southern blot analysis as contiguous DNA fragments. They were subcloned into pBluescript and partially sequenced to determine the exon-intron organization. As shown in Fig. 3B, the gene consists of a small first exon and a large second exon. The exon-intron borders conform to the GT/AG rule.
It may be interesting to note that the exon-intron organization of RIG-G is also highly conserved during evolution, because RIG-G, human ISG 54K, C.L ISG54K, and two murine ISGs, Ifi54k (for interferon-induced) and Ifi56k, genes are all composed of a small first exon and the second exon that contains all ORF except the ATG start codon and two nucleotides of the second codon (9, 12, 13) as shown in Fig. 3B.
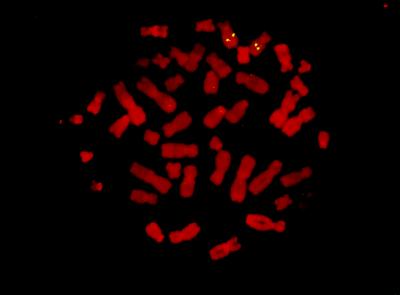
To determine the chromosomal locus of the RIG-G, two biotinylated probes prepared from the genomic clones were hybridized to metaphase chromosome of human lymphocytes. The hybridization with either of the probes showed signals on chromosome 10q24 (Fig. 6). Using PCR and Southern analysis, RIG-G, ISG-54K, and ISG-56K were clustered within a YAC contig of approximately 1 Mb (data not shown).
Figure 6.
The chromosome localization of RIG-G gene was determined by fluorescence in situ hybridization. Two spots of hybridization signals are present on both chromosome 10 at 10q24.
DISCUSSION
Because great success was achieved in the treatment of APL with ATRA (14), the APL cell line NB4 carrying chromosome translocation t(15;17) was selected and used as a model system to study the mechanism of ATRA-induced differentiation (15). Using the DD-PCR technique, we have identified the RIG-G gene, whose expression was induced at 12–24 hr after ATRA treatment. The fact that the protein synthesis inhibitor cycloheximide can completely block the ATRA induction of RIG-G expression suggests that ATRA-induced protein(s) must be synthesized to trigger the RIG-G expression, in contrast to ATRA-induced primary response genes such as RARα2 isoform. On the other hand, the expression of RIG-G can be rapidly induced by IFN-α in NB4 cells (30 min to 3 hr) and by IFN-α or IFN-γ in breast cancer cell lines.
It was previously reported that interaction of IFN with its receptor results in the induction of a group of ISGs (16). The expression of ISGs, such as human ISG54K, ISG56K, and C.L. ISG54K with cis-acting sequences IFN stimulation response element and IFN-γ activation sequence in promoter regions has been shown to be directly regulated at the transcriptional level through the Janus kinase–signal transducers and activators of transcription (STAT) pathway (12, 13, 17–19). Interestingly, RIG-G bears high homology with several ISGs in human (ISG54K, ISG56K, and IFN-inducible and retinoic acid-inducible 58K gene) and in other species. These genes should belong to the same family. In support of the view that RIG-G is a new member of this ISG family, our preliminary finding showed that the promotor region of RIG-G gene contains one relatively well conserved IFN-stimulation response element and several putative γ activation sequences (unpublished work). It is thus possible that RIG-G may be activated through the Janus kinase–STAT pathway.
Four mouse ISGs, namely Ifi54k, Ifi56k, Ifi56-ps1, and Ifi56-ps2, all could be assigned to the mouse 19D1 locus (13). The fluorescence in situ hybridization mapping in the present work determined that RIG-G gene was located on human chromosome 10q24, the same region where human ISG54K and ISG56K are located. The PCR and hybridization analysis of YAC contig confirmed that the three genes were clustered in a range of about 1 Mb. In addition, both primary sequences and exon-intron organization are well conserved among the related mouse and human genes. Therefore, they should come from the same origin by duplication during evolution.
The amino acid motifs deduced from the RIG-G cDNA sequence suggest that it could be the target molecule of phosphorylation by a number of important kinases, such as cAMP and cGMP-dependent kinase, casein kinase II, protein kinase C, and tyrosine kinase. RIG-G may thus be involved in many kinase pathways. The cytoplasmic localization of RIG-G protein is in good agreement with its possible role in cytosolic signal transduction. Tyrosine phosphorylation is the most likely way for RIG-G-related family members to participate in a signal transduction network because they all contain a consensus tyrosine phosphorylation motif, which is found at residue 109 in RIG-G. Although the precise function of RIG-G and its downstream partners remains to be identified, it seems that the expression of RIG-G may be involved in differentiation and anti-proliferation in ATRA-induced differentiation of APL cells. The fact that two other members of the ISG family, ISG54K (data not shown) and IFN-inducible and retinoic acid-inducible 58K gene reported by T. Niikura et al. (GenBank accession no. HSU34605) also were induced by ATRA suggests that ATRA could induce several members of this ISG family in NB4 cells, perhaps through a common pathway. Like RIG-G, other members of this family may also somehow participate in ATRA-induced differentiation. Besides, significant RIG-G induction was observed only in NB4 cells, but not in ATRA-susceptible HL-60 cells and breast cancer cell line, suggesting that this induction may need regulatory factors specific to APL cells.
It has been reported that retinoic acid and IFN can exert synergistic effects on regulating proliferation and differentiation in several tumor cell lines and in some clinical settings (20–22). Some studies suggested that these effects might be in part due to the synergy in inducing the expression of some genes, such as ISGs (23). Now our results show that ATRA and IFNs have synergistic effect on the transcription of a family of ISG, including RIG-G in NB4 cells (Fig. 4B). It is worth noting that our previously described RIG-E gene (6) is also regulated by both IFNs and ATRA in NB4 cells. These data are in line with our recent finding that IFN-γ potentiate the effect of ATRA in inducing the differentiation of ATRA-sensitive NB4 cells (data not shown) and the previous reports that combined use of ATRA and IFN could synergize in the induction of differentiation of fresh AML cells (24) and of maturation of APL cells resistant to ATRA (25). There is also a synergistic induction of the cyclin-dependent kinase inhibitor p21WAF1 by IFN-γ and ATRA in NB4 cells (26), which is followed by an increased G1 cell cycle arrest and induction of differentiation (27).
The mechanism of the synergism between ATRA and IFNs may involve STAT proteins, which are rapidly activated by IFN-induced phosphorylation. Among the six STATs (STAT 1α, 1β, 2, 3, 4, and 5) studied (28–31), only STAT1 was found to be up-regulated by ATRA-treated NB4 cells after 48 hr (ref. 32; unpublished work). Hence, the quantitative increment of STAT1 mRNA and proteins does not seem to be the primary event in the induced expression of RIG-G by ATRA. The dependence on protein synthesis for RIG-G expression may involve some proteins necessary for the post-translational phosphorylation of STAT-1 (32). ATRA also can interact with IFN signaling by regulating the positive and negative cofactors IRF-1 and IRF-2 (33). On the other hand, IFNs also can participate in the retinoid signaling pathway by regulating the expression of cytoplasmic and nuclear binding proteins (34). Understanding the network between ATRA and IFN signaling will facilitate the combinative use of these two agents in cancer therapy.
Acknowledgments
We thank Prof. J. E. Darnell of The Rockefeller University, New York for providing the ISG 54k and ISG 56k cDNA and Prof. G. Acs of Mount Sinai Medical University, New York, for critical review of the manuscript. This work is in part supported by grants from the Chinese Climbing Project, the National Natural Science Foundation of China, the European Community–China Biotechnology Program, and the Shanghai Life Science Center. M.Y., L.-X.K. and Y.-W.S. are supported by the Samuel Waxman Cancer Research Foundation and the C. Wu Foundation of the Shanghai Institute of Hematology. S.W. received National Institutes of Health Grant RO1 CA59936–04.
ABBREVIATIONS
- APL
acute promyelocytic leukemia
- PML
promyelocytic leukemia
- ATRA
all-trans-retinoic acid
- IFN
interferon
- RAR
retinoic acid receptor
- DD-PCR
differential display–PCR
- RIG-G
retinoic acid-induced gene G
- ISG
interferon-stimulated gene
- STAT
signal transducers and activators of transcription
- YAC
yeast artificial chromosome
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U52513).
References
- 1.Sachs L. Nature (London) 1978;274:535–539. doi: 10.1038/274535a0. [DOI] [PubMed] [Google Scholar]
- 2.de The H, Chomienne C, Lanotte M, Degos L, Dejean A. Nature (London) 1990;347:558–561. doi: 10.1038/347558a0. [DOI] [PubMed] [Google Scholar]
- 3.Chen Z, Tong J-H, Dong S, Zhu J, Wang Z-Y, Chen S-J. Gene Chromosomes Cancer. 1996;15:147–156. doi: 10.1002/(SICI)1098-2264(199603)15:3<147::AID-GCC1>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida H, Kitamura K, Tanaka K, Omura S, Miyazaki T, Hachiya T, Ohno R, Naoe T. Cancer Res. 1996;56:2945–2948. [PubMed] [Google Scholar]
- 5.Raelson J V, Nervi C, Rosenauer A, Benedetti L, Monczak Y, Pearson M, Pelicci P G, Miller W H., Jr Blood. 1996;88:2826–2832. [PubMed] [Google Scholar]
- 6.Mao M, Yu M, Tong J-H, Ye J, Zhu J, Huang Q-H, Fu G, Yu L, Zhao S-Y, Waxman S, Lanotte M, Wang Z-Y, Chen S-J, Chen Z. Proc Natl Acad Sci USA. 1996;93:5910–5914. doi: 10.1073/pnas.93.12.5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Licht J D, Shaknovich R, English M A, Melnick A, Li J Y, Reddy J C, Dong S, Chen S-J, Zelent A, Waxman S. Oncogene. 1996;12:323–336. [PubMed] [Google Scholar]
- 8.Cherif D, Julier C, Delattre O, Derre J, Lathrop G M, Berger R. Proc Natl Acad Sci USA. 1990;87:6639–6643. doi: 10.1073/pnas.87.17.6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy D E, Larner A C, Chaudhuri A, Babiss L E, Darnell J E., Jr Proc Natl Acad Sci USA. 1986;83:8929–8933. doi: 10.1073/pnas.83.23.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wathelet M G, Moutschen S, Defilippi P, Cravador A, Collet M, Huez G A, Content J. Eur J Biochem. 1986;155:11–17. doi: 10.1111/j.1432-1033.1986.tb09452.x. [DOI] [PubMed] [Google Scholar]
- 11.Lee C G L, Demarquoy J, Jackson M J, O’Brien W E. J Immunol. 1994;152:5758–5767. [PubMed] [Google Scholar]
- 12.Bluyssen H A R, Vlietstra R J, van der Made A, Trapman J. Eur J Biochem. 1994;220:395–402. doi: 10.1111/j.1432-1033.1994.tb18636.x. [DOI] [PubMed] [Google Scholar]
- 13.Bluyssen H A R, Vlietstra R J, Faber P W, Smit E M, Hagemeijer A, Trapman J. Genomics. 1994;24:137–148. doi: 10.1006/geno.1994.1591. [DOI] [PubMed] [Google Scholar]
- 14.Chen S-J, Wang Z-Y, Chen Z. Stem Cells. 1995;13:22–31. doi: 10.1002/stem.5530130104. [DOI] [PubMed] [Google Scholar]
- 15.Lanotte M, Martin-Thouvenin V, Najman S, Ballerini P, Valensi F, Berger R. Blood. 1991;77:1080–1086. [PubMed] [Google Scholar]
- 16.Friedman R L, Manly S P, McMahon M, Kerr I M, Stark G R. Cell. 1984;38:745–755. doi: 10.1016/0092-8674(84)90270-8. [DOI] [PubMed] [Google Scholar]
- 17.Larner A C, Jonak G, Cheng Y-S E, Korant B, Knight E, Darnell J E., Jr Proc Natl Acad Sci USA. 1984;81:6733–6737. doi: 10.1073/pnas.81.21.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larner A C, Chaudhuri A, Darnell J E., Jr J Biol Chem. 1986;261:453–459. [PubMed] [Google Scholar]
- 19.Darnell J E, Jr, Kerr I M, Stark G R. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 20.Peck R, Ballog W, Richard P, Werner B. Eur J Cancer. 1991;27:53–57. doi: 10.1016/0277-5379(91)90061-h. [DOI] [PubMed] [Google Scholar]
- 21.Hemmi H, Breitman T R. Blood. 1987;69:501–507. [PubMed] [Google Scholar]
- 22.Marth C, Daxenbichler G, Dapunt O. J Natl Cancer Inst. 1986;77:1197–1202. [PubMed] [Google Scholar]
- 23.Moore D M, Kalvakolane D V, Lippman S M, Kavanagh J J, Hong W K, Borden E C, Krakoff I H. Semin Hematol. 1994;31:31–37. [PubMed] [Google Scholar]
- 24.Gallagher R E, Lurie K J, Leavitt R D, Wiernik P H. Leukemia Res. 1987;11:609–619. doi: 10.1016/0145-2126(87)90033-6. [DOI] [PubMed] [Google Scholar]
- 25.Nason-Burchenal K, Gandini D, Botto M, Allopenna J, Seale J R C, Cross N C P, Goldman J M, Dmitrovsky E, Pandolfi P P. Blood. 1996;88:3926–3936. [PubMed] [Google Scholar]
- 26.Waxman S, Jing Y-K, Chen S-J, Chen Z. Blood. 1996;88:69a. (abstr.). [Google Scholar]
- 27.Manfredi J J, Tang H Y, Waxman S. Mol Cell Differ. 1996;4:33–45. [Google Scholar]
- 28.Yamamot K, Quelle F W, Thierfelder W E, Kreider B L, Gilbert D J, Jenkins N A, Copeland N G, Silvennoinen O, Ihle J N. Mol Cell Biol. 1994;14:4342–4349. doi: 10.1128/mcb.14.7.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gouilleux F, Wakao H, Mundt M, Groner B. EMBO J. 1994;13:4361–4369. doi: 10.1002/j.1460-2075.1994.tb06756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong Z, Wen Z, Darnell J E., Jr Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 31.Fu X-Y, Schindler C, Improta T, Aebersold R, Darnell J E., Jr Proc Natl Acad Sci USA. 1992;89:7840–7843. doi: 10.1073/pnas.89.16.7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gianni M, Terao M, Fortino I, LiCalzi M, Viggiano V, Barbui T, Rambaldi A, Garattini E. Blood. 1997;89:1001–1012. [PubMed] [Google Scholar]
- 33.Matikainen S, Ronni T, Hurme M, Pine R, Julkunen I. Blood. 1996;88:114–123. [PubMed] [Google Scholar]
- 34.Widschwendter M, Daxenbichler G, Bachmair F, Muller E, Zeimet A G, Uhl S M, Lang T, Marth C. Anticancer Res. 1996;16:369–374. [PubMed] [Google Scholar]