Abstract
The ultra-long telomeres that have been observed in mice are not in accordance with the concept that critical telomere shortening is related to aging and immortalization. Here, we have used quantitative fluorescence in situ hybridization to estimate (T2AG3)n lengths of individual telomeres in various mouse strains. Telomere lengths were very heterogeneous, but specific chromosomes of bone marrow cells and skin fibroblasts from individual mice had similar telomere lengths. We estimate that the shortest telomeres are around 10 kb in length, indicating that each mouse cell has a few telomeres with (T2AG3)n lengths within the range of human telomeres. These short telomeres may be critical in limiting the replicative potential of murine cells.
Telomeres, the physical ends of chromosomes, have a structure that is important for the stability and integrity of chromosomes (1, 2). Telomeres of vertebrate organisms end in (T2AG3)n repeat sequences of variable length (3). Telomere shortening results from incomplete end replication and may limit the replicative potential of various cells (4, 5). In humans, a critical shortening of telomeres has been related to cell senescence and oncogenesis (refs. 6 and 7; reviewed in ref. 8). In mice, however, ultra-long (T2AG3)n repeat sequences have been described, and shortening has not been observed upon aging (9, 10) or oncogenesis (11). These observations in the mouse have challenged the concept of telomere shortening in relation to aging and immortalization (8, 12–14).
Telomere lengths are measured with Southern blot analysis of terminal restriction fragments (TRFs) after DNA digestion with frequently cleaving restriction enzymes (3, 4, 8–10). This analysis results in a smear of TRFs, ranging from 5 to 15 kb in humans and from 20 to 150 kb in mice. Primarily, TRF heterogeneity will result from true variations in the number of T2AG3 repeats of individual telomeres. These variations in telomeres may be intercellular, interchromosomal, or intrachromosomal. It was demonstrated that in yeast cells heterogeneity is generated in the length of individual telomeres following multiple cell divisions (15). Also, in a human cell line with a telomere-associated marker sequence, variation in the length of this specific telomere was observed upon multiple cell divisions (16). However, alternative mechanisms may also contribute to the heterogeneity of TRF lengths. There may be a variable distance from restriction enzyme site to the (T2AG3)n sequence in various telomeres. Also, some TRFs may be derived from intersitial rather than terminal (T2AG3)n sequences (17). Clearly, these issues cannot be addressed by TRF analysis since it requires DNA derived from >105 cells.
Here, we have used quantitative fluorescence in situ hybridization (FISH) to estimate the (T2AG3)n length of individual mouse telomeres. Overall, telomeres were very heterogeneous, but specific chromosomes of bone marrow cells and skin fibroblasts had a similar telomere length, indicating that telomere heterogeneity results mainly from heritable interchromosomal variations.
MATERIALS AND METHODS
Mice.
BALB/c, C57BL/6, C3H, 129, and DBA/2 mice were obtained from The Jackson Laboratories and were bred and maintained in the animal facility of the British Columbia Cancer Research Centre (Vancouver). Mice were killed at the age of 8–12 weeks for harvests of cells.
Bone Marrow Cells.
Bone marrow cells were obtained by flushing both femora with Hanks’ balanced salt solution supplemented with 2% fetal calf serum using a syringe and a fine needle. Cells were cultured in serum-free medium as described (18), supplemented with human interleukin 6 (10 ng/ml; Immunex), human granulocyte colony-stimulating factor (10 ng/ml; Amgen Biologicals), human erythropoietin (3 units/ml; Stem Cell Technologies, Vancouver), murine interleukin 3 (20 ng/ml from supernatants of COS cells transfected with a murine interleukin 3 cDNA), and murine SF (steel factor or mast cell growth factor) (50 ng/ml; Immunex). Colcemid (0.05 mg/ml) was added for 2 h prior to cell harvest on day 3.
Skin Fibroblasts.
Skin biopsies (10 × 10 mm) were cut in 1 × 1 mm pieces and placed in Petri dishes in DMEM with 20% fetal calf serum. After 4–5 days, fibroblasts were growing and the skin explants were removed. Colcemid (0.05 mg/ml) was added for 6 h on day 10–14 of culture.
In Situ Hybridization.
Bone marrow cells and skin fibroblasts were harvested from cultures following colcemid incubation. After hypotonic swelling in KCl buffer (30 min at 37°C), cells were fixed and stored in methanol/acetic acid according to standard procedures (19). Before hybridization, cells were dropped on slides and dried overnight. After washing with PBS, slides were fixed in formaldehyde (4%) in PBS (2 min), washed again with PBS (three times for 5 min) and treated with pepsin (P-7000; Sigma) at 1 mg/ml at pH 2 (10 min at 37°C). Formaldehyde fixation and washing steps were repeated and slides were dehydrated in ethanol and air dried. Hybridization mixture (2 × 15 μl), containing 70% formamide, 0.3 μg/ml Cy-3-conjugated (C3TA2)3 peptide nucleic acid (PNA) probe (PBIO/Biosearch Product, Bedford, MA), 0.25% (wt/vol) blocking reagent (DuPont) in 10 mM Tris (pH 7) was added to the slide, a coverslip (22 × 60 mm) was added followed by DNA denaturation (3 min at 80°C). After hybridization for 2 h at room temperature, slides were washed with 70% formamide/10 mM Tris (pH 7.2) (two times for 15 min) and with 0.05 M Tris/0.15 M NaCl (pH 7.5) containing 0.05% Tween-20 (three times for 5 min). Slides were dehydrated with ethanol, air dried, and covered by 2 × 5 μl antifade solution (Vectashield; Vector Laboratories) containing 0.2 μg/ml of 4′-6-diamidino-2-phenylindole (DAPI).
Quantitative Image Analysis.
Digital images were recorded with a MicroImager MI1400–12 camera (Xillix; Vancouver) on an Axioplan fluorescence microscope (Zeiss) equipped at the excitation site with a multiple filter wheel (Pacific Scientific, Rockford, IL), a DAPI/Cy-3 dual band emitter filter at the emission site (Chroma Technology, Brattleboro, VT), and a focusing motor drive (ZSS 43-200-1.2; Phyptron, Grödenzell, Germany) and controller (MAC4000; Marzhauser, Wetzlar, Germany). Microscope control and image acquisition was performed with dedicated software (ssm; Xillix). Images were acquired with a Plan-Apochromat ×63, NA 1.4 objective lens (Zeiss) and a mercury/xenon lamp (200 W; Optiquip, Highland Mills, NY). A dedicated computer program was developed for image analysis. In short, chromosomes and telomeres were identified through segmentation of the DAPI image and the Cy-3 image, respectively. Both images were combined and corrected for pixel shifts. The integrated fluorescence intensity for each telomere was calculated after correction for background, based on the values of the surrounding pixels, and after correction for image acquisition exposure time. Finally, the integrated fluorescence intensity of individual telomeres is expressed in a table for each chromosome which can be subjected to editing (see Fig. 1C). To prevent a possible selection bias, images were acquired “blindly”—i.e., metaphases were chosen solely on the basis of a good chromosome spread in the DAPI image and without looking at the corresponding Cy-3 image. All acquired images were included in the analysis.
Figure 1.
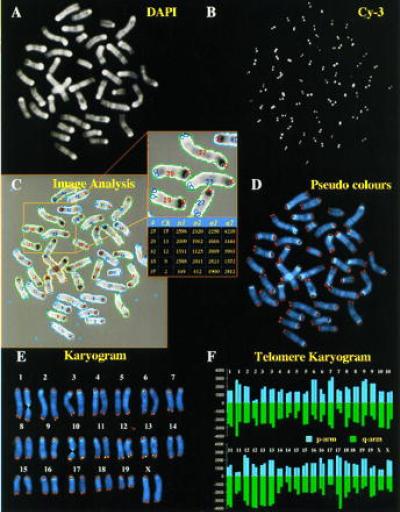
Quantitative FISH analysis of individual telomeres illustrated on a bone marrow-derived metaphase spread from a BALB/c mouse. (A) Digital image of DAPI-stained chromosomes. (B) Individual telomeres in the Cy-3 fluorescence image. (C) Output of dedicated image analysis software which combines the DAPI and Cy-3 images, identifies chromosomes and telomeres, assigns colors to the four telomeres (purple, red, light and dark blue), calculates the integrated fluorescence intensity for each telomere after correction for background and expresses the integrated fluorescence intensity of individual telomeres of each chromosome in a table. (D) Combination of DAPI and Cy-3 images after assigning pseudocolors—i.e., blue for DAPI and orange for Cy-3, with the use of Adobe photoshop (Adobe Systems, Mountain View, CA). (E) Individual chromosomes after karyotyping according to standard DAPI banding patterns (19). (F) Finally, the integrated fluorescence intensity is expressed for both p- and q-arm telomeres from each karyotyped chromosome.
Calibration.
We used two levels of calibration to ensure a reliable quantitative estimation of telomere length in various samples. First, to correct for daily variations in lamp intensity and alignment, images of fluorescent beads (orange beads, size 0.2 μm; Molecular Probes) were acquired and similarly analyzed with the image analysis computer program. Second, to relate fluorescence intensity to number of T2AG3 repeats, we hybridized and analyzed plasmids with a defined (T2AG3)n length of 0.15, 0.40, 0.80, and 1.60 kb (20). There was a linear correlation (r = 0.99) for plasmid fluorescence intensity and (T2AG3)n length with a slope of 43.1. Therefore, the calibration corrected telomere fluorescence intensity (ccTFI) of each telomere was calculated according to the formula: ccTFI = (Bea1/Bea2) × (TFI/43.1), where Bea1 = fluorescence intensity of beads when plasmids were analyzed, Bea2 = fluorescence intensity of beads when sample x was analyzed, and TFI = unmodified fluorescence intensity of a telomere in sample x.
A restriction of this calibration method is that the actual telomeres are outside the range of the (T2AG3)n length of the plasmids. The assumption is made that the linear correlation obtained between fluorescence intensity and telomere insert size in plasmid DNA is maintained in the higher range.
Statistical Analysis.
Results are expressed in histograms or box plots. The horizontal lines in the box plots express the 5th, 25th, 50th, 75th, and 95th percentile of the distribution. Differences between groups were analyzed with the Wilcoxon rank sum test. Sample size estimates were calculated according to the formula: n/group = 2× [(Zα + Zβ) × σ/Δ]2 for a significance level of 0.05 (α = 0.95 and Zα = 1.64) and a power of 80% (β = 0.90 and Zβ = 1.28), where n = number, σ = standard deviation, and Δ = difference between the two groups.
RESULTS
Quantitative FISH.
To investigate the length of the telomeric (T2AG3)n repeat sequence at the end of individual chromosomes, we have used quantitative FISH (21). Chromosome preparations of bone marrow cells and skin fibroblasts were hybridized with a synthetic 18-mer PNA probe with a (CCCTAA)3 sequence. The probe was directly conjugated with the fluorescent dye Cy-3. Quantitative FISH analysis was enabled by using the PNA probe at conditions that favor hybridization of probe but prevent renaturation of telomere repeat sequences (21, 22). Digital images were acquired from DAPI-stained chromosomes (Fig. 1A) and from Cy-3-stained telomeres (Fig. 1B). Contours were made from individual chromosomes and individual telomere spots with the use of a dedicated image analysis computer program. Individual chromosomes were combined with the corresponding four telomere spots (Fig. 1C). The integrated light intensity of each telomere spot was calculated and expressed as arbitrary fluorescence units for telomeres p1,p2 for short arm telomeres and q1,q2 for long arm telomeres (table in Fig. 1C). The two images were also combined after assigning pseudocolors—i.e., blue to DAPI and orange to Cy-3 (Fig. 1D) and a karyogram was made (Fig. 1E). Finally, the fluorescence intensity of p- and q-arm telomeres on all individual chromosomes was expressed in arbitrary units (Fig. 1F).
Telomere Lengths in Inbred Mouse Strains.
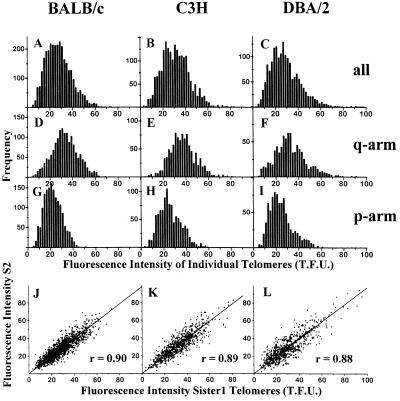
The fluorescence intensity of the various telomeres in an individual mouse cell is very heterogeneous (Fig. 1 B and D). Following quantitative analysis, this heterogeneity is demonstrated for three mice from BALB/c, C3H, and DBA/2 strains, respectively (Fig. 2 A–C). Fluorescence intensity of individual telomeres was related to length of the (T2AG3)n repeat sequence using a number of calibration steps (see Materials and Methods). Telomere fluorescence intensity of individual telomeres (x-axis of Figs. 2 and 3 and y-axis of Fig. 4) are expressed so that 1 telomere fluorescence unit corresponds to 1 kb of (T2AG3)n repeats. Our results suggest that telomere lengths in these mice are in the range from 5–10 to 50–60 kb, occasionally up to 80 kb (Fig. 2 A–C). Similar telomere length distributions were observed in all other mice that we analyzed from BALB/c (n = 9), 129 (n = 2), C3H (n = 2), C57BL/6 (n = 8), and DBA/2 (n = 2) strains.
Figure 2.
Telomere fluorescence intensities in three mice, from BALB/c, C3H, and DBA/2 strains respectively. Histograms express the fluorescence intensity and frequency of all individual telomeres from 12 to 15 bone marrow-derived metaphases (A–C). Individual telomeres on the distal q-arm (D–F), and proximal p-arm (G–I) are expressed separately. The differences in median fluorescence intensity between p- and q-arm telomeres were highly significant (P < 0.0001, Wilcoxon rank sum test) in all three mice. Dot plots (J–L) express the fluorescence intensity values of the two sister chromatids (S1 and S2) on each chromosome end. Correlation coefficients (r) varied from 0.88 to 0.90. Telomere fluorescence intensity is expressed so that 1 telomere fluorescence unit (T.F.U.) corresponds to 1 kb of (T2AG3)n sequence according to results from similarly hybridized and analyzed plasmids with a defined (T2AG3)n length.
Figure 3.
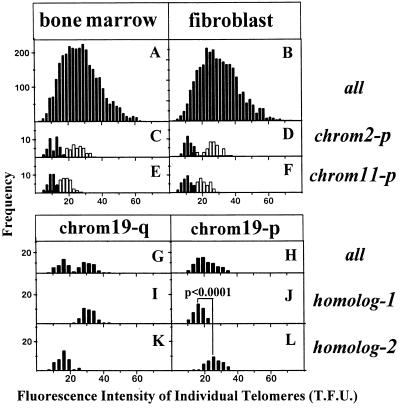
Short telomeres are present on specific chromosomes and variability in telomere length on homologous chromosomes. Histograms express the fluorescence intensity of individual telomeres from 15 bone marrow- (A) or skin fibroblast-derived (B) metaphases of a single BALB/c mouse. The telomeres on chromosome 2-p (C and D) and 11-p (E and F) are separated in two subpopulations: one derived from homolog with short telomeres (filled bars) and one from the corresponding homolog with longer telomeres (open bars) in each cell. (Lower) Telomeres from chromosome 19-q (G) and 19-p (H) are shown for the bone marrow-derived cells. The 19-q telomeres show a histogram with bimodal distribution originating from one homolog with a longer telomere (I) and one with a shorter telomere (K) in each cell. When the two homologs of chromosome 19 are distinguished according to this difference in 19-q telomeres, there is also a highly significant, reversed difference for p-arm telomeres (J and L). One telomere fluorescence unit (T.F.U.) corresponds to 1 kb of (T2AG3)n length (see also the legend to Fig. 2).
Figure 4.
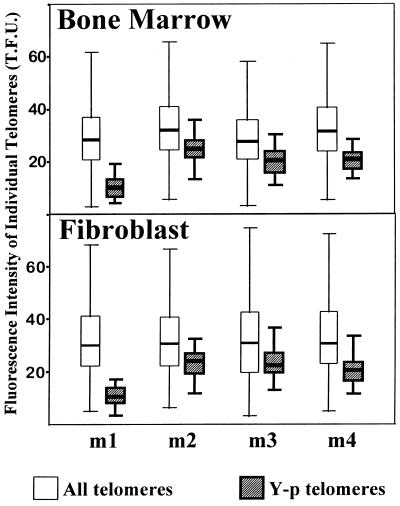
Heterogeneity of Y-chromosome telomeres in four male BALB/c littermates (m1–m4). The open box plots represent all telomeres from 15 bone marrow cells or skin fibroblasts. The hatched box plots represent only the telomeres of the proximal p-arm of the Y-chromosome in the same cell populations. The median telomere fluorescence values for total telomeres varied from 28.4 to 32.0 in the bone marrow and fibroblast populations from the various mice. The median telomere values of Y-p telomeres in each mouse in bone marrow and skin fibroblast, respectively, are as follows: m1, 10.1 and 10.6; m2, 25.0 and 24.0; m3, 20.7 and 22.4; m4, 21.0 and 21.2. One telomere fluorescence unit (T.F.U.) corresponds to 1 kb of (T2AG3)n length (see also the legend to Fig. 2).
Correlation Between Telomeres on Sister Chromatids.
The fluorescence intensities of the two sister chromatids on each metaphase chromosome were highly correlated (Fig. 2 J–L), indicating that a low fluorescence value for one telomere was associated with a low value for the telomere on the related sister chromatid. This phenomenon can also be seen directly in the telomere fluorescence image (Fig. 1). The two related sister chromatids (p1 and p2 or q1 and q2) of individual chromosomes are likely to have identical or almost identical telomeres since one is copied from the other in the preceding S phase. Although the overall correlation is high, some sister chromatid telomeres show variations of almost 2-fold (Fig. 2 J–L). In this variation, quantitative FISH method-related variability may be an important contribution (see Discussion).
Telomeres Are Shorter on the p-Arms than on the q-Arms of Mouse Chromosomes.
All mouse chromosomes, except the Y-chromosome, are acrocentric (19). Probably, mouse chromosomes are truly telocentric because euchromatic DNA with coding genes has not been identified between centromeres and p-arm telomeres (23). We observed significantly higher (P < 0.0001, Wilcoxon rank sum) fluorescence intensities of q-arm telomeres (Fig. 2 D–F) in comparison to p-arm telomeres (Fig. 2 G–I) in all mice from BALB/c, 129, C3H, C57BL/6, and DBA/2 strains. Also, q-arm telomeres were found to have more length heterogeneity than p-arm telomeres (Fig. 2 D–I), resulting in a comparable length of the shortest telomeres on p-arms and q-arms. These results suggest that mechanisms for telomere elongation act differently on p-arm and q-arm telomeres. The proximity of p-arm telomeres to centromeric heterochromatin may affect binding of telomerase or telomerase-regulating proteins. Alternatively, telomere elongation through recombination may occur less frequently on p-arm telomeres.
Similar Telomere Length of Specific Chromosomes in Somatic Cells.
We observed that in individual mice, specific chromosomes are characterized by a specific telomere length. In the bone marrow-derived metaphase of the BALB/c mouse represented in Fig. 1, the p-telomeres of one of the homologs of chromosomes 2 and 11 have relatively short telomeres (Fig. 1 E and F). Short telomeres were found on one of the homologs of the same chromosomes in all bone marrow cells (Fig. 3 C and E) and skin fibroblasts (Fig. 3 D and F) in this mouse. In other mice, from identical or different strains, the shortest telomeres were often found on other chromosomes—e.g., 3-q, 5-p, 18-p, or X-p. The difference in the telomere length of homolog chromosomes is illustrated for chromosome 19 of the same BALB/c mouse (Fig. 3 G–L). The 19-q telomeres in this mouse showed a bimodal distribution (Fig. 3G) that was related to one homolog with a shorter telomere (Fig. 3I) and one with a longer telomere (Fig. 3K) in each cell. When the two homologs of chromosome 19 in each cell were distinguished according to this difference in q-telomeres, there was also a highly significant reversed difference for the telomeres on the corresponding p-arms (Fig. 3 J and L). This indicates that the distinction of the two homolog chromosomes on the basis of 19-q telomere length does indeed identify two different subpopulations. The presence of one 19-homolog with longer p-arm telomeres and one with longer q-arm telomeres in this mouse can also be seen in Fig. 1E. An identical difference between the two homologs of chromosome 19 was observed in skin fibroblasts in this mouse (data not shown). The observation that identical chromosomes in unrelated tissues from an individual mouse have a comparable telomere length suggests that the telomere length of each chromosome is determined very early in fetal development and that limited heterogeneity is generated during somatic cell divisions. However, the observation that homologous chromosomes in mice from inbred strains have different telomere lengths also suggests that substantial heterogeneity in telomere length is generated in germ line cells.
Generation of Telomere Heterogeneity in Germ Line.
To test the hypothesis that telomere length heterogeneity is generated in the germ line, a family of four male BALB/c mice was investigated (Fig. 4). In the first littermate (m1) the shortest telomere was found on the p-arm of the Y-chromosome with a length of around 10 kb in bone marrow and fibroblasts. In contrast, the other littermates have Y-p telomeres of 20–25 kb. Because all mice have inherited this Y-chromosome from the same father, these results indicate that considerable heterogeneity—i.e., from 10 to 25 kb—was generated in the paternal germ-line cells. Unfortunately, cells from the father were not available for study. However, the family analysis suggests that it was 20–25 kb because this length was found in three of the four littermates. It is unlikely that a reduction to 10 kb—i.e., the Y-p telomere in littermate m1—resulted solely from an end-replication-related loss of 75 bp per cell division (24). More likely, this reduction in Y-p telomere length was the result of (unequal) recombination (25–28) or rapid telomere deletion (29).
DISCUSSION
Here, we have used quantitative FISH to measure the number of T2AG3 repeat sequences at the end of individual murine chromosomes. In individual mice, telomeres were very heterogeneous but specific chromosomes of bone marrow cells and skin fibroblasts had a similar telomere length. This observation indicates that telomere length is heritable among somatic cells. However, homologous chromosomes in these inbred mice were often characterized by widely different telomere length, indicating that telomere heterogeneity is frequently generated in the germ line.
The results of this study provide additional support for the use of quantitative FISH to estimate the (T2AG3)n length of individual telomeres (21). However, there are several potential limitations related to the quantitative FISH method that may result in inaccuracies of the estimated number of telomeric T2AG3 repeats. These include inadequate accessibility of target sequences for the PNA probe, DNA loss during denaturation (30), inadequate focusing, and unequal illumination of the microscope field. Theoretically, all these mechanisms will result in random and not chromosome-specific, decreases in hybridization signals. Several observations argue against a major role for such random processes. First, the analysis of metaphase chromosomes allows each telomere to be measured in duplicate because the telomere on one sister chromatid has been copied from the other in the preceding S phase. We observed a high correlation of telomeres fluorescence values on the related sister chromatids (r = 0.85–0.95). Second, the observation that the shortest telomeres in all cells from an individual mouse were found on a specific chromosome strongly argues against such a random process. Theoretically, there might still be a chromosome-specific reduced accessibility or increased DNA loss. However, the family studies show that specific chromosomes (e.g., the Y-chromosome) can have hybridization signals corresponding to 10- or 25-kb telomeres in various littermates, arguing against such a chromosome-specific reduction in hybridization signal. Nevertheless, chromosome-specific telomeres (Figs. 3 and 4) and pairs of related sister telomeres (Fig. 2) showed maximal variations of ≈2-fold. These variations were random, and methodological inaccuracies may have been important contributions here. The presence of such variations underscores the necessity to analyze more than one metaphase spread to obtain useful estimates of (average) telomere length. This consideration limits the applicability of quantitative FISH in studies of telomere dynamics. Nevertheless, sample size estimates indicate that a 3-kb shortening of a chromosome-specific telomere of 10 kb (typically measured with a standard deviation of 2.5–3.0 kb) requires the analysis of only 10 metaphases.
Our estimates of telomere lengths using quantitative FISH are shorter than previous estimates using TRF analysis (9, 10). In these studies, mice from DBA/2, CBA, and BALB/c strains were found to have TRFs ranging from 20 to 150 kb, and C57BL/6 strains ranging from 20 to 60 kb. We observed essentially identical telomere lengths of 10–60 kb, occasionally up to 80 kb, in all mice from BALB/c, DBA/2, 129, C3H, and C57BL/6 strains. It has been suggested that the ultra-long TRFs in mice (9, 10) are derived from interstitial (T2AG3)n sequences rather than from terminal, telomeric sequences. We have not observed interstitial chromosome staining using this sensitive PNA–FISH in any of the strains tested although the presence of such sites close to the actual telomere (within several megabase) would be difficult to resolve from the telomere hybridization signal. Most likely, the difference in telomere length estimation between TRF and quantitative FISH analysis results from an overestimation of actual (T2AG3)n length with TRF analysis related to the physical distance between restriction enzyme site and terminal (T2AG3)n repeats (reviewed in ref. 8). The subtelomeric regions of yeast (31) and humans (32) are composed of various repeat sequences that may result in restriction enzyme sites that are not directly adjacent to the terminal (T2AG3)n repeat sequence. This is of particular importance in mice, since the proximal telomere is closely linked to the centromere. TRFs of >1 Mb, containing centromeric minor satellite DNA, were found after digestion with restriction enzymes that have a 6-bp recognition sequence (23). Our data are compatible with 10–100 kb of non-T2AG3 sequences contributing to murine TRFs that are derived from digestion of genomic DNA with frequently cleaving enzymes such as MboI (9–11). Our results also suggest that the 60- to 150-kb TRF polymorphism observed between strains and individual mice results from polymorphism in subtelomeric restriction enzyme site rather than from polymorphism in telomere length.
The observation that in individual mice specific chromosomes are characterized by a similar telomere length in tissues as different as bone marrow cells and skin fibroblasts indicates that the telomere length of individual chromosomes is a heritable factor that is maintained throughout the somatic cell divisions in mouse development. Most likely, the telomere length that is present on each chromosome in the parental spermatocyte or oocyte is a major determinant of telomere length in somatic cells. In view of this observation, the degree of telomere length diversity that is generated on specific chromosomes in the germ line of the mouse is surprising. Assuming that there is a decrease in telomere length of 75–100 bp per cell division, up to a few kilobases of heterogeneity in the length of telomeres of specific chromosomes in different germ cells is expected. Yet, variations of up to 15–20 kb were found for telomeres of homologous chromosomes within a highly inbred mouse colony and for telomeres on specific chromosomes between littermates. It is unlikely that an increased telomerase activity accounts for the larger variability of specific telomeres in germ cells. Unlike in humans, telomerase is expressed in mice not only in testis but also in somatic cells (24). Probably, telomere length variation in germ-line cells is specifically related to the pairing and recombination of homolog chromosomes during meiosis (33), similar to the unequal recombination between homolog chromosomes that has been observed for other repetitive DNA sequences (25).
In accordance with the results of TRF analysis (9, 10), we observed that the overall length of telomeres is longer in mice than in humans. However, the heterogeneity of chromosome-specific telomeres is also larger in mice than in humans—i.e., ≈6-fold (this study) and ≈3-fold, respectively (U.M.M., J.M.J.M.Z., S.S.S.P., V. Dragowska, J. Yui, E. A. Chavez, R.K.W., and P.M.L., unpublished data). This is in agreement with the observation that the shortest telomeres in the mouse are within the range of human telomeres. It has been demonstrated that fibroblasts of Mus spretus lose ±75 bp of telomeres in vitro with each cell division (24), similar to the rate of telomere loss in human fibroblasts (4). Using quantitative FISH, we found a highly significant 4-kb shortening of telomeres in BALB/c-derived bone marrow cells that were induced to maximal proliferation with serial bone marrow transplantation (J.M.J.M.Z., U.M.M., A.K.R., H.J.T., and P.M.L., unpublished data). Telomere length reduction was observed not only for overall telomeres but also for the shortest. Therefore, it can be hypothesized that the shortest telomeres in a cell rather than its overall length may be rate-limiting in its proliferative capacity. Indeed, studies in yeast have shown that loss of a single telomere may result in cell cycle arrest (2, 34). Quantitative FISH, with the capacity to investigate telomeres of specific chromosomes, will be indispensable in testing this hypothesis. In particular, it appears of interest to examine telomere shortening on specific chromosomes in murine cells upon aging or oncogenesis as such shortening was difficult to demonstrate for TRFs with an average length of 30 kb (10, 11).
Acknowledgments
We thank E. Chavez, N. Verwoerd, and J. Wiegant for technical assistance; T. de Lange (The Rockefeller University) for providing plasmids and helpful discussions; V. Ling and B. Palcic (BC Cancer Research Centre) for making equipment available for these studies; and StemCell, Immunex, and Amgen for reagents. This work was supported by grants from the Dutch Cancer Society (J.M.J.M.Z.) and the Deutsche Forschungsgemeinschaft (U.M.M.), and grants from the National Institutes of Health and the National Cancer Institute of Canada with funds from the Terry Fox Run.
ABBREVIATIONS
- TRF
terminal restriction fragments
- FISH
fluorescence in situ hybridization
- PNA
peptide nucleic acid
- DAPI
4′-6-diamidino-2-phenylindole
References
- 1.Blackburn E H. Cell. 1994;77:621–623. doi: 10.1016/0092-8674(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 2.Zakian V A. Science. 1995;270:1601–1607. doi: 10.1126/science.270.5242.1601. [DOI] [PubMed] [Google Scholar]
- 3.Moyzis R K, Buckingham J M, Cram L S, Dani M, Deaven L L, Jones M D, Meyne J, Ratliff R L, Wu J-R. Proc Natl Acad Sci USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allsopp R C, Vaziri H, Patterson C, Goldstein S, Younglai E V, Futcher A B, Greider C W, Harley C B. Proc Natl Acad Sci USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaziri H, Dragowska W, Allsopp R C, Thomas T E, Harley C B, Lansdorp P M. Proc Natl Acad Sci USA. 1994;91:9857–9860. doi: 10.1073/pnas.91.21.9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hastie N D, Dempster M, Dunlop M G, Thompson A M, Green D K, Allshire R C. Nature (London) 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 7.Harley C B, Futcher A B, Greider C W. Nature (London) 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 8.de Lange T. In: Telomere Dynamics and Genome Instability in Human Cancer. Blackburn E H, Greidere C W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 265–293. [Google Scholar]
- 9.Starling J A, Maule J, Hastie N D, Allshire R C. Nucleic Acids Res. 1990;18:6881–6888. doi: 10.1093/nar/18.23.6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kipling D, Cooke H J. Nature (London) 1990;347:347–402. doi: 10.1038/347400a0. [DOI] [PubMed] [Google Scholar]
- 11.Broccoli D, Godley L A, Donehower L A, Varmus H E, de Lange T. Mol Cell Biol. 1996;16:3765–3772. doi: 10.1128/mcb.16.7.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harley C B, Kim N W, Prowse K R, Weinrich S L, Hirsch K S, West M D, Bacchetti S, Hirte H W, Counter C M, Greider C W, Wright W E, Shay J W. Telomerase, Cell Immortality, and Cancer. Vol. 59. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. pp. 307–315. [DOI] [PubMed] [Google Scholar]
- 13.Autexier C, Greider C W. Trends Biochem Sci. 1996;21:387–391. [PubMed] [Google Scholar]
- 14.Lundblad V, Wright W E. Cell. 1996;87:369–375. doi: 10.1016/s0092-8674(00)81358-6. [DOI] [PubMed] [Google Scholar]
- 15.Shampay J, Blackburn E H. Proc Natl Acad Sci USA. 1988;85:534–538. doi: 10.1073/pnas.85.2.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murnane J P, Sabatier L, Marder B A, Morgan W F. EMBO J. 1994;13:4953–4962. doi: 10.1002/j.1460-2075.1994.tb06822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kipling D, Salido E C, Shapiro L J, Cooke H J. Nat Genet. 1996;13:78–82. doi: 10.1038/ng0596-78. [DOI] [PubMed] [Google Scholar]
- 18.Lansdorp P M, Dragowska W. J Exp Med. 1992;175:1501–1509. doi: 10.1084/jem.175.6.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J J, Warburton D, Robertson E J. Anal Biochem. 1990;189:1–17. doi: 10.1016/0003-2697(90)90036-9. [DOI] [PubMed] [Google Scholar]
- 20.Hanish J P, Yanowitz J L, de Lange T. Proc Natl Acad Sci USA. 1994;91:8861–8865. doi: 10.1073/pnas.91.19.8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lansdorp P M, Verwoerd N P, van de Rijke F M, Dragowska V, Little M-T, Dirks R W, Raap A K, Tanke H J. Hum Mol Genet. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- 22.Egholm M, Buchardt O, Christensen L, Behrens C, Freier S, Driver D A, Berg R H, Kim S K, Norden B, Nielsen P E. Nature (London) 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 23.Kipling D, Ackford H E, Mitchell A R, Taylor B A, Cooke H J. Genomics. 1991;11:235–241. doi: 10.1016/0888-7543(91)90128-2. [DOI] [PubMed] [Google Scholar]
- 24.Prowse K R, Greider C W. Proc Natl Acad Sci USA. 1995;92:4818–4822. doi: 10.1073/pnas.92.11.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harbers K, Soriano P, Mueller U, Jaenisch R. Nature (London) 1986;324:682–685. doi: 10.1038/324682a0. [DOI] [PubMed] [Google Scholar]
- 26.Biessmann H, Mason J M. Adv Genet. 1992;30:185–249. doi: 10.1016/s0065-2660(08)60321-1. [DOI] [PubMed] [Google Scholar]
- 27.Wang S-S, Zakian V A. Nature (London) 1990;345:456–458. doi: 10.1038/345456a0. [DOI] [PubMed] [Google Scholar]
- 28.McEachern M J, Blackburn E H. Genes Dev. 1996;10:1822–1834. doi: 10.1101/gad.10.14.1822. [DOI] [PubMed] [Google Scholar]
- 29.Li B, Lustig A J. Genes Dev. 1996;10:1310–1326. doi: 10.1101/gad.10.11.1310. [DOI] [PubMed] [Google Scholar]
- 30.Raap A K, Marijnen J G H, Vrolijk J, van der Ploeg M. Cytometry. 1986;7:235–242. doi: 10.1002/cyto.990070303. [DOI] [PubMed] [Google Scholar]
- 31.Chan C S M, Tye B-K. Cell. 1983;33:563–573. doi: 10.1016/0092-8674(83)90437-3. [DOI] [PubMed] [Google Scholar]
- 32.Brown W R A, MacKinnon P J, Villasante A, Spurr N, Buckle V J, Dobson M J. Cell. 1990;63:119–132. doi: 10.1016/0092-8674(90)90293-n. [DOI] [PubMed] [Google Scholar]
- 33.Sen D, Gilbert W. Nature (London) 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 34.Sandell L L, Zakian V A. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]