Abstract
Background
Sperm cells must regulate the timing and location of activation to maximize the likelihood of fertilization. Sperm from most species, including the nematode Caenorhabditis elegans, activate upon encountering an external signal. Activation for C. elegans sperm occurs as spermatids undergo spermiogenesis, a profound cellular reorganization that produces a pseudopod. Spermiogenesis is initiated by an activation signal that is transduced through a series of gene products. It is now clear that an inhibitory pathway also operates in spermatids, preventing their premature progression to spermatozoa and resulting in fine-scale control over the timing of activation. Here, we describe the involvement of a newly assigned member of the inhibitory pathway: spe-4, a homolog of the human presenilin gene PS1. The spe-4(hc196) allele investigated here was isolated as a suppressor of sterility of mutations in the spermiogenesis signal transduction gene spe-27.
Results
Through mapping, complementation tests, DNA sequencing, and transformation rescue, we determined that allele hc196 is a mutation in the spe-4 gene. Our data show that spe-4(hc196) is a bypass suppressor that eliminates the need for the spermiogenesis signal transduction. On its own, spe-4(hc196) has a recessive, temperature sensitive spermatogenesis-defective phenotype, with mutants exhibiting (i) defective spermatocytes, (ii) defective spermatids, (iii) premature spermatid activation, and (iv) spermatozoa defective in fertilization, in addition to a small number of functional sperm which appear normal microscopically.
Conclusion
A fraction of the sperm from spe-4(hc196) mutant males progress directly to functional spermatozoa without the need for an activation signal, suggesting that spe-4 plays a role in preventing spermatid activation. Another fraction of spermatozoa from spe-4(hc196) mutants are defective in fertilization. Therefore, prematurely activated spermatozoa may have several defects: we show that they may be defective in fertilization, and earlier work showed that they obstruct sperm transfer from males at mating. hc196 is a hypomorphic allele of spe-4, and its newly-discovered role inhibiting spermiogenesis may involve known proteolytic and/or calcium regulatory aspects of presenilin function, or it may involve yet-to-be discovered functions.
Background
Sperm cells can be considered as semi-autonomous organisms having their own unique genomes and living lives that are, in part, independent of the organisms that produced them. Sperm are exceedingly variable across animal taxa in terms of morphology, motility, and lifespan. One seemingly invariant feature is that a sperm cell's active life must initiate with precise timing. Precocious activation may result in premature consumption of stored energy or interfere with efficient fertilization. Belated activation may allow competing sperm to gain the advantage in the race to fertilize the ova. In many cases, activation coincides with sperm release from the male. For example, in sea urchins, a longstanding model for sperm function, Na+ enters the sperm when they are released into seawater. A simultaneous pH spike stimulates motility and respiration through a signal transduction pathway that includes spikes in Ca2+, cAMP, and cAMP dependent kinases. The result is the activation of dynein ATPase in the flagellum and increased mitochondrial activity [1-3]. The sudden ionic flux that accompanies release into seawater is the cue that initiates sperm motility in sea urchins.
In internal fertilizing species, the sperm also require a cue to initiate activity. The cue is often a signaling molecule. In many mammals, bicarbonate serves as the cue, inducing a Ca2+ influx, cAMP accumulation, cAMP dependent protein kinase A activity, and increased motility and respiration [4-7]. Thus, even though sea urchins and mammals are distantly related and the signals for sperm activation are different, some of the cellular processes involved in sperm activation are similar.
Here, we are concerned with the cellular mechanisms that regulate sperm activation in the nematode Caenorhabditis elegans. Like all nematode sperm, those from C. elegans are amoeboid, and attainment of activity coincides with the developmental transition from spermatid to spermatozoon (= spermiogenesis). In hermaphrodites, spermiogenesis occurs as the spermatids enter the sperm storage chamber, or spermatheca. In males, spermiogenesis occurs during copulation as the spermatids encounter seminal fluid [8]. In both cases, the timing of spermiogenesis is critical. For instance, belated activation may result in the spermatids being swept from the reproductive tract by passing eggs [9], whereas prematurely activated spermatozoa within the male reproductive tract impede sperm transfer [10]. The signaling molecule that initiates spermiogenesis in C. elegans is unknown, but in a manner similar to the species discussed above, sperm activation in C. elegans appears to involve the influx of cations [11], a brief elevation in pH [12], and the release of intracellular Ca2+ [8,13,14].
The control of C. elegans spermiogenesis involves a number of gene products. The signal to activate is transduced through a pathway that includes the genes spe-8, spe-12 [15,16], spe-19 [17], spe-27 [18], and spe-29 [19]. Mutations in these genes result in arrest at the spermatid stage in hermaphrodite produced self-sperm. Curiously, mutant males are fertile, and the arrested hermaphrodite produced self-sperm can be "rescued" when exposed to seminal fluid from males, which, along with other data, suggests that this "spe-8" group of genes amplifies the activation signal for hermaphrodite self sperm [8,16]. In vitro, the arrest associated with mutations in the spe-8 group genes can be overcome by inducing one of the events that occurs downstream of the signal transduction pathway: elevation of pH. This is accomplished by treating the cells with the weak base triethanolamine [12]. Thus, C. elegans sperm possess a complex spermiogenesis pathway that ensures the timely activation of spermatids.
It is now clear that spermatid activation in C. elegans also involves negative regulation. The propensity to undergo spermiogenesis is so great that an inhibitory pathway is required to prevent premature progression through this important developmental transition. This inhibitory pathway includes spe-6. A large screen for mutations that suppress the sterile phenotype of spe-27(it132ts) identified a number of mutations in spe-6, which result in premature spermatid activation and in suppression of mutations of the spe-8 group genes [20]. SPE-6 inhibits spermiogenesis until the activation signal is transduced through the spe-8 group gene products. SPE-6 also functions earlier in spermatogenesis in the packaging of Major Sperm Protein (MSP) into the fibrous body-membranous organelles (FB-MOs), which are unique to the sperm cells [21]. Here, we report on the inhibitory function of SPE-4 on spermatid activation. spe-4, a homolog of the human presenilin gene PS1, had been known for its role in MSP packaging in the FB-MOs early in spermatogenesis [21-23]. Our investigation focuses on a hypomorphic spe-4 allele isolated in the same suppressor screen that produced the spe-6 suppressor alleles. Like the spe-6 suppressor alleles, spe-4(hc196) suppresses mutations in the spe-8 group genes and results in premature spermatid activation.
Results and Discussion
Genetic mapping places hc196 right of center on chromosome I
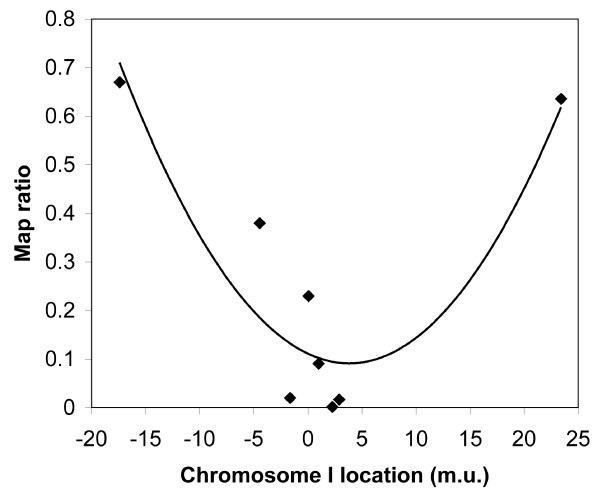
We initially mapped hc196 to chromosome I using single nucleotide polymorphisms from the polymorphic Hawaiian strain CB4856. Linkage to chromosome I was unequivocal (map ratio to SNP on chromosome: I – 0.00, II – 1.01, III – 1.63, V – 1.58; ratios near zero indicate linkage [24]). We then undertook mapping with SNP loci across chromosome I. The calculated map ratios for each of the SNP loci analyzed are shown in Fig. 1. The second order equation for the best fit curve (y = 0.0014 × 2 - 0.0105x + 0.1111) has a minimum at 3.75, which is a rough estimate of the locus. Based upon the fact that the SNP loci at 2.26 (pkP1117) and 2.88 (pkP1119) showed almost no recombination with the Hawaiian chromosomes, we chose to conduct a three factor mapping cross with markers that flank this region: dpy-5 and unc-29, located at 0.00 and 3.32, respectively. The cross was performed in a spe-27 background, so the hc196 mutant phenotype was fertility. We recovered 34 recombinant F2 progeny: 19 recombination events occurred between dpy-5 and hc196 (5 fertile dpy and 14 sterile unc), and 15 recombination events occurred between hc196 and unc-29 (14 sterile dpy and 1 fertile unc). Based upon these data, hc196 resides at 1.9 m.u. on chromosome I (19/34 × 3.32).
Figure 1.
Single nucleotide polymorphism mapping results. The map ratio represents the ratio of N2 to CB4658 DNA from the bulk isolated DNA from mutant and wild-type worms. A map ratio near one indicates no linkage, whereas map ratios near zero are found for loci nearby the mapped gene [24]. The curve is the best-fit second order equation: y = 0.0014 × 2 - 0.0105x + 0.1111.
hc196 is an allele of spe-4
Due to its proximity to spe-4 at 2.04 on chromosome I, we decided to perform a complementation test to determine if hc196 is an allele of spe-4. Worms with the hc196/spe-4(hc78) genotype were sterile (Table 1), indicating that hc196 is an allele of spe-4. There are six other alleles of spe-4, and all are sterile (only eb27, a nonsense mutation, produces one offspring per 100 mutant worms [22]). The fact that hc196 homozygotes are partially fertile suggests that the allele is hypomorphic and that these worms have some SPE-4 function. Interestingly, the trans-heterozygote, composed of a hypomorphic allele over a null allele (hc196/hc78), was sterile (Table 1). Apparently, SPE-4 activity from only a single hypomorphic allele is too little to allow functional sperm production.
Table 1.
Results of a complementation test between hc196 and spe-4(hc78).
| Hermaphrodite Genotype | Brood Size ± SEM (N) |
| hc196 +/spe-4(hc78) unc-15; spe-27/+ | 0.6 ± 0.3 (62) |
| spe-4(hc78) unc-15 | 0 ± 0 (15) |
| hc196 | 16.6 ± 1.8 (27) |
| spe-4(hc78) unc-15/++ | 95.3 ± 5.7 (12) |
| hc196/+ | 94.8 ± 14.1 (9) |
| spe-27/+ | 199.3 ± 11.8 (12) |
Worms were reared at 25°C.
The identity of hc196 as an allele of spe-4 was supported by transformation rescue experiments. N2 worms transformed with myo-2::GFP and a PCR product containing wild-type spe-4 and its flanking DNA (zqEx1 [myo-2::GFP spe-4(+)]; see Methods) were mated to dpy-5 hc196; spe-27 unc-22 hermaphrodites. We scored the fertility of F2 worms expressing GFP (to select for the transgenes) and a dumpy phenotype (to select for hc196 homozygotes) but lacking an uncoordinated phenotype (to avoid the spe-27 sterile phenotype). The dumpy transformants had significantly larger brood sizes than did non-transgenic dumpy animals (Table 2; t = -5.38, P < 0.001), showing that the wild-type spe-4 transgene rescued the hc196 fertility defect.
Table 2.
Brood size of worms in transformation rescue experiments.
| Hermaphrodite Genotype | Brood Size ± SEM (N) |
| dpy-5 hc196; spe-27 unc-22 | 8.8 ± 1.0 (9) |
| dpy-5 hc196/++; spe-27 unc-22/++ | 63.4 ± 6.1 (7) |
| dpy-5 hc196 | 14.9 ± 1.0 (11) |
| Transgenic worms* | 53.5 ± 5.7 (15) |
* These were dpy-5 hc196; spe-27 unc-22/++ or dpy-5 hc196; ++ worms transformed with the transgene zqEx1 [myo-2::GFP spe-4(+)]. Worms were reared at 25°C.
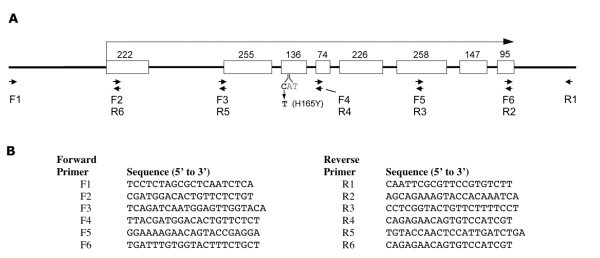
Finally, we sequenced spe-4 from our hc196 strain and identified a missense mutation. Every base pair was covered by at least two sequencing reads from a cloned region of hc196 DNA containing spe-4. The mutation was confirmed by two additional sequencing reads on an independent PCR product amplified with primers F3 and R4 (Fig. 2). The mutation alters a histidine residue that is conserved in every nematode SPE-4 homolog known (Fig. 3). spe-4 is an ortholog of the human presenilin gene PS1, but a paralog two other PS1 orthologs in the C. elegans genome: sel-12 and hop-1. The SPE-4 amino acid position affected by the hc196 mutation is conserved in HOP-1 but not SEL-12. Our mutation lies in a membrane spanning domain, and the majority of the mutations isolated in SPE-4 (all of which are apparent null mutations) lie within 35 amino acids of hc196 (Fig. 3). Indeed, the hc78 missense mutation is nearby in the same transmembrane domain, suggesting that this domain is important to SPE-4 function, even though the catalytic aspartates involved in the proteolytic Presenilin-1 function are located downstream (Fig. 3).
Figure 2.
Molecular analysis of spe-4. A. Structure of spe-4. The boxes represent exons with the number of base pairs above. The hc196 mutation is shown below the third exon. The arrows represent oligonucleotides used for PCR amplification and DNA sequencing. B. The sequence of the oligonucleotides.
Figure 3.
Alignment of nematode SPE-4 protein sequences. C. elegans SPE-4 is aligned with SPE-4 sequences from several nematode species and with C. elegans SEL-12 and HOP-1, the other presiniln orthologs in the C. elegans genome. Positions that are shaded and boxed represent identities, whereas boxed positions represent chemical similarity. The hc196 mutation is indicated above the altered amino acid, as are the other mutations in C. elegans SPE-4. The blue lines beneath the sequences represent transmembrane domains as predicted for C. elegans SPE-4 by TMPred (Release: TMBase25) [42]. The positions with yellow shading represent the catalytic aspartates involved in Presenilin 1 proteolysis as predicted by the ELM Server (Release: May 2007) [27]. Accession numbers for the protein sequences are: C. elegans SPE-4, C. briggsae SPE-4, CAE66611.1; Brugia malayi SPE-4, EDP33786.1; C. elegans SEL-12, NP_508175.1; and C. elegans HOP-1, NP_491328.1. The C. remanei SPE-4 sequence was obtained from WormBase.org, accession number RP:RP21351 (Release WS184).
In an attempt to uncover additional spe-4 suppressor alleles, we investigated a collection of spe-27 suppressor mutants isolated in the same mutagenesis that generated spe-4(hc196) [25]. Because spe-4(hc196) males produce very few cross progeny, we mated heterozygous males [spe-4(hc196)/+; spe-27] to hermaphrodites of the other suppressor strains (which were all in a spe-27 unc-22 background). The F1 were isolated and kept at 25°C. None of the additional alleles tested resulted in fertile F1 progeny (Table 3). The fact that all the additional alleles complement spe-4(hc196) indicates that none of the additional suppressor mutations tested were suppressor alleles of spe-4. As controls, we tested three spe-6 alleles that suppress the sterility of spe-27 [25], and all of them complemented hc196 as well. The absence of additional alleles is curious, given the fact that 18 suppressor alleles of spe-6 were isolated in the spe-27 suppression mutagenesis that produced spe-4(hc196) [20]. At 1398 base pairs, the coding sequence of spe-4 is a bigger target for mutation than that of spe-6 at 1140 base pairs, so we expected to find multiple spe-4 suppressor alleles. Perhaps spe-6 plays a more central role in the inhibitory pathway or spe-4 has fewer amino acids critical to the inhibitory pathway function.
Table 3.
Results of complementation tests between spe-4(hc196) and additional spe-27 suppressor alleles.
| Suppressor Allele | Brood size ± SEM (N) | Results |
| hc166 | 0.5 ± 0.2 (25) | Complementation |
| hc176 | 0.4 ± 0.2 (16) | Complementation |
| hc197 | 0.5 ± 0.1 (21) | Complementation |
| hc198 | * | Complementation |
| hc199 | 0.1 ± 0.1 (15) | Complementation |
| hc201 | 0.1 ± 0.1 (8) | Complementation |
| hc202 | * | Complementation |
| hc204 | 0.4 ± 0.3 (10) | Complementation |
| hc205 | 1.2 ± 0.6 (5) | Complementation |
| zq6 | 0 ± 0 (20) | Complementation |
| zq7 | 0.3 ± 0.2 (13) | Complementation |
| zq8 | 0.2 ± 0.1 (16) | Complementation |
| spe-6(hc163) | 0.6 ± 0.2 (10) | Complementation |
| spe-6(hc187) | 0.4 ± 0.2 (10) | Complementation |
| spe-6(hc188) | 0.6 ± 0.2 (22) | Complementation |
The brood sizes were measured in a spe-27(it132ts) background at 25°C.
* Complementation tests for these alleles were conducted by Paul Muhlrad [25]
spe-4(hc196) is a recessive bypass suppressor of spermiogenesis pathway mutations and has a conditional Spe phenotype on its own
Worms homozygous for spe-27(it132ts) are sterile at 25°C, unless they are also homozygous for spe-4(hc196) (see Table 4). This suppression of spe-27 sterility is a recessive phenotype, as spe-4(hc196)/+ are sterile in a spe-27 background. In an otherwise wild-type background, spe-4(hc196) worms have reduced fertility that is influenced by temperature: they produce five times more progeny at 20°C than they do at 25°C (Table 4). This fertility deficit is a due to a sperm defect, because the fertility of spe-4(hc196) hermaphrodites increases after males supply sperm at mating (Table 4). Furthermore, the spe-4(hc196) phenotype is recessive; spe-4(hc196)/+; spe-27/+ worms have wild-type fertility (Table 4).
Table 4.
Brood sizes of various strains. All spe-27 alleles were it132ts
| Temperature-dependent Brood Size ± SEM (N) | |||
| Hermaphrodite Genotype | 15C | 20C | 25C |
| spe-4(hc196); spe-27 unc-22 | 43.0 ± 7.2 (17) | 64.0 ± 6.9 (21) | 11.0 ± 0.7 (64) |
| spe-4(hc196); spe-27 | 59.8 ± 9.1 (13) | 100.4 ± 8.6 (14) | 14.6 ± 2.1 (14) |
| spe-27 unc-22 | 103.6 ± 11.7 (5) | 4.2 ± 1.0 (4) | 0.2 ± 0.2 (5) |
| spe-27 | 236.2 ± 17.2 (5) | 2.0 ± 0.5 (5) | 0.2 ± 0.1 (63) |
| spe-4(hc196)/+; spe-27 | 0.2 ± 0.1 (30) | ||
| spe-4(hc196)/+; spe-27/+ | 191.0 ± 19.0 (11) | ||
| spe-27/+ | 199.3 ± 11.8 (12) | ||
| spe-4(hc196) | 84.8 ± 2.8 (5) | 84.8 ± 4.8 (5) | 16.6 ± 1.8 (27) |
| spe-4(hc196) × N2 males | 135.2 ± 6.1 (4) | 123.8 ± 15.3 (5) | |
| dpy-5(e61) spe-4(hc196); spe-8(hc134ts) | 18.6 ± 1.6 (9) | ||
| spe-8(hc134ts) | 0.3 ± 0.2 (10) | ||
| dpy-5(e61) spe-4(hc196); spe-19(eb52) | 15.2 ± 1.4 (19) | ||
| spe-19(eb52) | 0 ± 0 (10) | ||
| spe-4(hc196); spe-29(it129) dpy-20(e1282ts) | 14.2 ± 1.9 (20) | ||
| spe-29(it129) dpy-20(e1282ts) | 0.1 ± 0.1 (10) | ||
| N2 | 303.6 ± 19.8 (5) | 246.6 ± 12.2 (5) | 190.8 ± 5.4 (5) |
Comparing the spe-4(hc196) suppression phenotype with its own phenotype in a wild-type spe-27 genetic background suggests that spe-4(hc196) is epistatic to spe-27. The fertility of the double mutant more closely resembles spe-4(hc196) than it does spe-27(it132ts) (Table 4). This argument assumes that the suppression and sterile phenotypes are brought about by the same mechanism. If they are due to different mechanisms, then the epistatic relationship between these genes is not obvious from our data.
We were interested in the mechanism by which spe-4(hc196) suppresses spe-27 sterility. To test whether the interaction between the two genes is specific, we assessed the ability of spe-4(hc196) to suppress the sterility of mutations in the spermiogenesis genes spe-8, spe-19, and spe-29 (a double spe-4 spe-12 mutant was not made because spe-12 is tightly linked to spe-4; the two genes are only ~0.6 map units apart). In each case, spe-4(hc196) restored fertility to the spermiogenesis gene mutants (Table 4). Therefore, the suppression phenotype of spe-4(hc196) is not allele or gene specific, suggesting that it bypasses the spermiogenesis pathway in a manner similar to the spe-6 suppressor alleles described by Muhlrad and Ward [20].
We also tested three putative null spe-4 alleles for suppression of spe-27. These three alleles produce a sterile phenotype. One is a missense mutation (hc78), while the other two are molecular nulls: hc81 (amber stop), and q347 (deletion) (Fig. 3). In each case, the spe-4; spe-27 double worms were sterile (Table 5). This result was not surprising, given that these spe-4 mutations cause spermatocyte arrest and the virtual absence of spermatids available to undergo spermiogenesis [22,23]. What was surprising was that the heterozygous state of each of the three alleles did suppress spe-27 (Table 5). The suppression of spe-27 by heterozygous spe-4 alleles that are null suggests that a simple reduction (but not elimination) of SPE-4 function is sufficient for suppression. It is interesting that suppression by spe-4(hc78)/+ resulted in larger broods than either of the two molecular nulls, suggesting that spe-4(hc78) is not a null allele, even though it induces complete sterility. Its residual function is only apparent in the suppression phenotype. Furthermore, the trans-heterozygote, dpy-5 spe-4(hc196) +/+ spe-4(hc78) unc-15; spe-27 unc-22 also suppressed spe-27 (Table 5), which is of interest because the trans-heterozygote was sterile when two both copies of the spe-27 gene were wild type (Table 1).
Table 5.
Suppression of spe-27 by different spe-4 alleles.
| Hermaphrodite Genotype | Brood Size ± SEM (N) |
| spe-4(hc78) | |
| spe-4(hc78) unc-15 | 0 ± 0 (32) |
| spe-4(hc78) unc-15/+ + | 141.0 ± 11.8 (6) |
| spe-4(hc78) unc-15; spe-27 unc-22 | 0.7 ± 0.3 (9) |
| spe-4(hc78) unc-15/++; spe-27 unc-22 | 26.7 ± 3.6 (31) |
| spe-4(q347) | |
| dpy-5(e61) spe-4(q347) | 0 ± 0 (9) |
| dpy-5(e61) spe-4(q347)/+ + | 154.0 ± 25.0 (10) |
| dpy-5(e61) spe-4(q347); spe-27 unc-22 | 0 ± 0 (7) |
| dpy-5(e61) spe-4(q347)/+ +; spe-27 unc-22 | 13.8 ± 6.0 (14) |
| spe-4(hc81) | |
| dpy-5(e61) spe-4(hc81) | 0 ± 0 (8) |
| dpy-5(e61) spe-4(hc81)/+ + | 172.3 ± 33.3 (10) |
| dpy-5(e61) spe-4(hc81); spe-27 unc-22 | 0 ± 0 (8) |
| dpy-5(e61) spe-4(hc81)/+ +; spe-27 unc-22 | 10.4 ± 4.4 (11) |
| spe-4(hc196)/spe-4(hc78) | |
| dpy-5 spe-4(hc196) +/+ spe-4(hc78) unc-15; spe-27 unc-22 | 15.3 ± 3.8 (3) |
Worms were reared at 25°C
spe-4(hc196) causes multiple sperm defects including premature spermiogenesis
Mutant spe-4(hc196) hermaphrodites produce very few progeny at 25°C (Table 4). We mated these hermaphrodites to fer-1(hc13ts); him-5 males to see if more of their sperm would become functional after exposure to seminal fluid from fer-1 males, whose own sperm are defective. Such is the case with spe-8 group mutant hermaphrodite sperm, which activate and become functional when exposed to male seminal fluid [8,15,18]. We examined seven spe-4(hc196) hermaphrodites that had been paired with sterile fer-1 males: they produced an average of 151.1 progeny. Therefore, some of the spermatids within spe-4(hc196) hermaphrodites are similar to those in spe-8 group mutant hermaphrodites: they cannot activate unless exposed to male seminal fluid.
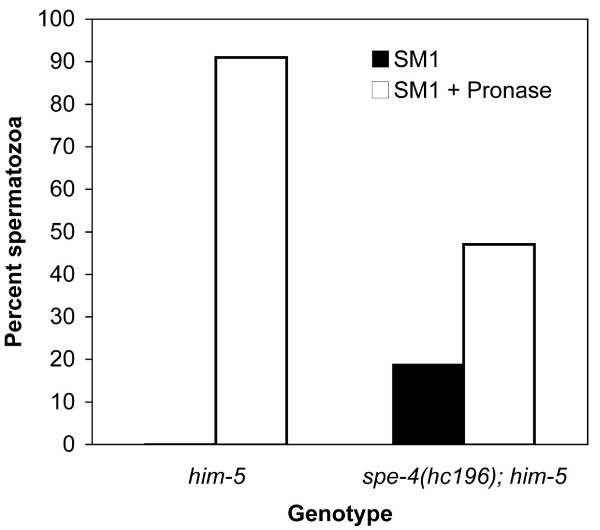
We also examined sperm from male spe-4(hc196); him-5 males. Compared to him-5 males, spe-4(hc196); him-5 male worms contained a large number of spermatocytes in the seminal vesicle where only spermatids should be found (CWL, personal observations), indicating that the hc196 mutation is an impediment to meiotic division, which is also a characteristic of the non-suppressor spe-4 mutations [22,23]. Some spermatocytes do successfully negotiate the meiotic divisions, and while mutant males accumulated spermatids, they also contained spermatozoa. In fact, nearly 20% of their sperm were crawling spermatozoa (Fig. 4). Typically, males store only spermatids, which undergo spermiogenesis after exposure to seminal fluid at ejaculation. The existence of spermatozoa within virgin males shows that they failed to pause in the spermatid stage. Furthermore, spe-4(hc196) males are nearly infertile, producing only small numbers of cross progeny even given 24 hours or more to mate. Such infertility may be due to premature spermatid activation because crawling spermatozoa impede sperm transfer during copulation [10], demonstrating the importance of the proper timing of sperm activation.
Figure 4.
Sperm activation in Pronase. The presence of spermatozoa in sperm released from males dissected in sperm medium (SM1) and in sperm medium containing the protease Pronase, which activates spermatids to spermatozoa in vitro.
In contrast, not all the spermatids stored within spe-4(hc196) males appear capable of undergoing spermiogenesis. The ability of sperm to undergo spermiogenesis can be examined in vitro using Pronase, a collection of proteases which apparently cleave one or more cell surface components and thereby induce spermiogenesis [12]. While more than 90% of the spermatids in him-5 males activate to spermatozoa in the presence of Pronase, only ~50% of the sperm from spe-4(hc196) males activated in Pronase exposure (Fig. 4). Because 20% of the sperm these males contained was activated prior to exposure, Pronase treatment only activated 30% of the sperm. Therefore, almost 50% of the mutant sperm failed to activate. However, we did not observe any sperm that appeared "stalled" in the spiky intermediate stage of spermiogenesis. Remaining as spiky intermediates is a characteristic of the sperm from spe-8 group mutants when exposed to Pronase [8]. Thus, sperm from spe-4(hc196) mutants share some characteristics with the spe-8 group mutants that they suppress, but not all their characteristics.
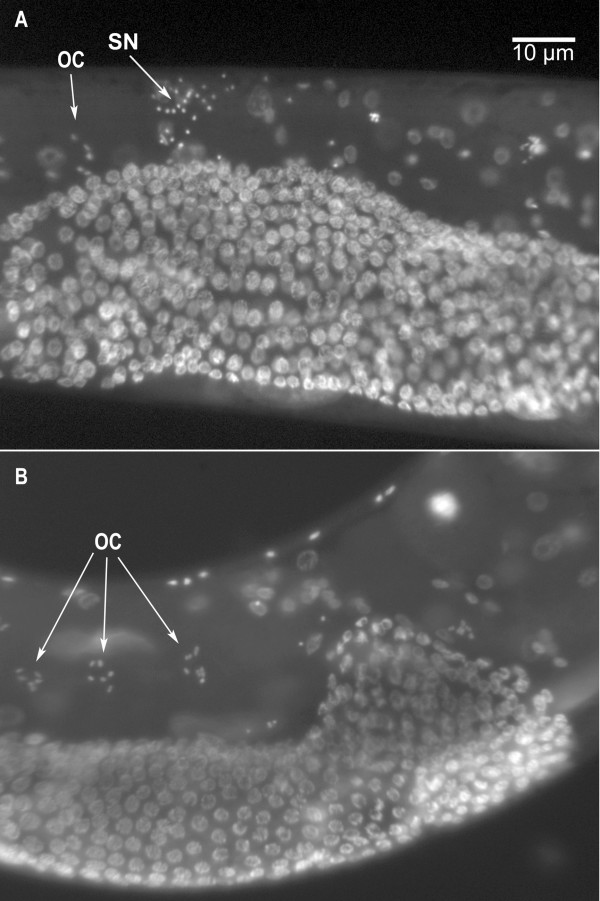
We had great difficulty obtaining cross progeny from spe-4(hc196); him-5 males, so we tested their fertility by pairing them with fer-1(hc13ts) hermaphrodites that had been raised at 25°C. The fer-1 mutation disrupts sperm function: fer-1 hermaphrodites are self-sterile and lose their defective self-sperm rapidly as they are swept from the reproductive tracts by passing unfertilized oocytes [26]. In 18 matings between individual spe-4(hc196); him-5 males each with four fer-1 unc-29 hermaphrodites, 13 males failed to sire any progeny, and together, the males averaged 1.22 progeny each (SD = 2.31). The males that sired no progeny actually did transfer sperm to the fer-1 unc-29 hermaphrodites, as evidenced by sperm within the mated hermaphrodites' spermathecae that were never found in unmated controls (Fig. 5). Therefore, at least some of the sperm from the mutant males were transferred during mating and crawled to the spermatheca, but they were unable to fertilize the oocytes.
Figure 5.
Sperm from spe-4(hc196) males within fer-1 hermaphrodites. Epifluorescence images of (A) a fer-1(hc13ts) hermaphrodite that had been mated to spe-4(hc196); him-5(e1490) males, and (B) an age-matched unmated fer-1(hc13ts) hermaphrodite showing the absence of sperm nuclei. SN indicates the characteristically compact DAPI labeled nuclei from the male's sperm [43], which appear localized to the spermatheca. OC indicates the oocyte chromosomes that are nearly the same size as sperm nuclei but occur in groups of 6 and are not as brightly fluorescent.
spe-4 is a member of an inhibitory pathway preventing premature spermiogenesis
Because reduction of SPE-4 function bypasses the spermiogenesis activation pathway, spe-4 appears to be the second member of a spermiogenesis inhibition pathway. This pathway prevents progression through spermiogenesis until relieved by the signal transduced through the activation pathway consisting, at least in part, of spe-8, spe-12, spe-19, spe-27, and spe-29 (Fig. 6). The founding member of the inhibitory pathway is spe-6, a casein kinase I that is proposed to maintain inhibition through phosphorylation [20]. SPE-4 has multiple potential CK1 phosphorylation sites as predicted by the ELM Server (Release: May 2007) [27], so SPE-4 could be a direct target of SPE-6 CK1 activity.
Figure 6.
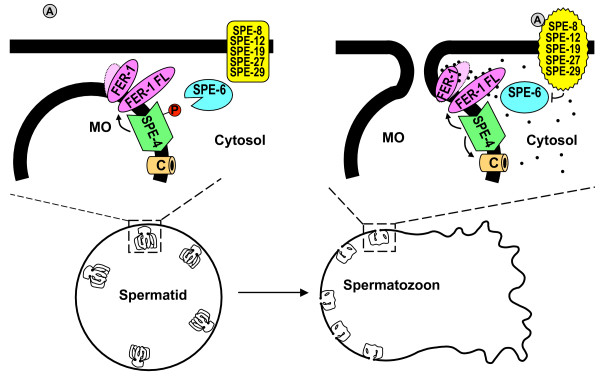
Hypothetical model of SPE-4 function in spermatid activation. In this model, SPE-6 phosphorylates downstream targets to maintain the spermatid stage [20]. We speculate that the Presenilin 1 homolog SPE-4 could be a target of SPE-6 kinase activity, increasing overall SPE-4 function. Based on the role of Presenilin 1 in cleaving amyloid precursor protein to produce two isoforms of amyloid β peptides, we further speculate that the potential proteolytic function of SPE-4 produces the two proteolytic isoforms of FER-1 found in sperm [14], although the target may be another protein. When the activation signal (A) is received and transduced by the spe-8 group proteins, SPE-6 function is suppressed, and SPE-4 function drops. There are at least two possible outcomes of reduced SPE-4 function: (i) a change in the ratio of the two proteolytic isoforms of FER-1 or another protein (denoted by the lighter and darker purple symbols for FER-1), which change in relation to spermiogenesis; and (ii) a spike in cytosolic Ca2+ (black dots) mediated through SPE-4 itself or through a calcium channel (C). The result is the FER-1 mediated fusion of the MOs to the plasma membrane and pseudopod formation.
The role of SPE-4 in the prevention of spermiogenesis may occur by several different mechanisms, but our results suggest that reduced SPE-4 function fosters precocious spermiogenesis. One possible mechanism involves the proteolytic function of the human PS1, a member of the γ-secretase complex whose members are located in the membranes of several cellular compartments including the Golgi. More than 150 mutations in PS1 contribute to early onset familial Alzheimer's Disease through abnormal deposition of the amyloid β-peptide (Aβ) in the brain [28,29]. Many of the PS1 mutations originally deemed to be gain of function are now thought to be loss of function lesions that decrease proteolytic efficiency but increase the ratio of the 42-residue to the 40-residue variant of Aβ [28,30]. We propose a model where the FER-1 protein is a target of SPE-4 proteolytic activity (Fig. 6). Spermatids contain two isoforms of FER-1, which likely result from proteolysis [14]. The human γ-secretase complex normally targets type I transmembrane proteins for proteolysis [31,32], but FER-1 is a type II membrane protein [33], casting doubt upon FER-1 as a target. However, we propose FER-1 as a SPE-4 target because (i) SPE-4 is the most divergent PS1 family member and may have different target specificities, (ii) both SPE-4 and FER-1 exist in the MO membrane, and (iii) FER-1 is cleaved during sperm maturation. In wild-type spermatids, activation likely happens when SPE-4 activity abates, perhaps as a result of decreasing SPE-6 function, resulting in an altered FER-1 isoform ratio that promotes spermiogenesis (Fig. 6). The hypothesis that FER-1 is a SPE-4 target requires further investigation.
Regulation of Ca2+ homeostasis is another potential PS1-like function of SPE-4 that may play a role in the inhibition of spermiogenesis. PS1 mutations have been shown to perturb Ca2+ homeostasis in human cells [34-36]. This perturbation may occur through a disruption in phosphatidylinositol 4,5-bisphosphate (PIP2) signaling [37]. PIP2 generally plays an important role in ion channel modulation [38]. Alternatively, SPE-4 might act as a Ca2+ leak channel itself [39]. If SPE-4 regulates Ca2+ homeostasis in spermatids either directly or through a PIP2-like intermediate, loss of function spe-4 mutations may affect sperm activation. SPE-4 resides in the MO membrane, the same location as TRP-3, a Ca2+ channel (Fig. 6) [40]. It is not clear if TRP-3 functions in spermiogenesis, but intracellular chelation of Ca2+ prevents spermiogenesis [14], indicating that spermiogenesis is Ca2+ dependent. This leads to the hypothesis that SPE-4 maintains Ca2+ sequestration in the spermatid phase and reduced SPE-4 activity allows Ca2+ entry into the cytosol (Fig. 6). The exact role of SPE-4 in the inhibition of spermiogenesis remains to be determined.
The inhibitory pathway that consists of spe-4 and spe-6 likely contains multiple other genes and involves the membranous organelles. The spe-27 suppression screen that generated the suppressor alleles of spe-4 and spe-6 produced at least four other mutated genes, two and perhaps more of which are not previously known spe genes (unpublished data). Both SPE-4 and FER-1 reside in the MOs [14,22]. The MOs exist in a complex with the fibrous bodies to form the FB-MO complexes that function to package the major sperm proteins (MSPs). MSP packaging is deficient for the likely null alleles of both spe-4 and spe-6 [21,22]. The FBs disassemble in the spermatid as the MSP is released, but the MOs remain and fuse with the plasma membrane during spermiogenesis [8]. Therefore, the MO is an important site of activity during spermiogenesis, and it is conceivable that the machinery of the spermiogenesis inhibition pathway resides in the MOs.
It appears that the spermiogenesis inhibition pathway is important for normal sperm function. Males with the spe-4(hc196) mutation were largely sterile, even though mutant hermaphrodites were partly fertile. The male form of sterility likely derives from several sources. One is the fact that many mutant spermatids do not activate (if our in vitro activation assay reflects what occurs in vivo). Therefore, even though some spermatids activate prematurely, others apparently cannot activate at all. Another likely source of sterility is the obstruction that active spermatozoa pose to sperm transfer [10]. Finally some mutant sperm exhibit a fertilization defect. Perhaps these spermatozoa underwent spermiogenesis so prematurely that they were not properly organized for fertilization. This suggests that the timing of sperm activation is not only important for interaction with the external environment, but also for correct assembly of the spermatozoon. In our studies with spe-4(hc196), premature activation results in only a small number of functional sperm; the majority are defective.
SPE-4 sequence divergence
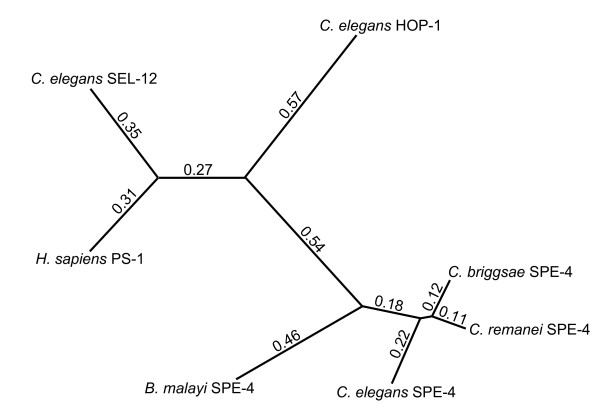
We performed an analysis of the similarities of the SPE-4 protein sequences from four nematodes along with those of SEL-12 and HOP-1, the two other C. elegans PS1 homologs, and human PS1 (Fig. 7). SEL-12 is the most similar of the nematode proteins to human PS1. HOP-1 is more diverged, and the four SPE-4 proteins cluster together. Therefore, it would appear that hop-1 and spe-4 are the result of duplication events since the separation of the human and nematode lineages. The spe-4 like genes are the most divergent of the PS1 orthologs, and because Brugia malayi has likely been separated from Caenorhabditis spp. for several hundred million years, the spe-4 type presenilins apparently have an ancient specialization that likely involves sperm function.
Figure 7.
Similarity relationships among selected nematode Presenilin-1 homologs. The similarities among the protein sequences were calculated using a Neighbor-Joining method with Poisson correction. The numbers and branch lengths represent relative sequence differentiation among the sequences. In addition to the sequences found in Fig. 3, the human Presenilin 1 sequence accession number is BAD96893.1.
Conclusion
spe-4 is the second member of a potentially new inhibitory pathway that prevents premature spermiogenesis in C. elegans. spe-4(hc196) was isolated in the same suppressor screen that produced multiple suppressor alleles of spe-6. This screen identified mutations that restored partial fertility to spe-27(it132ts) hermaphrodites. The suppressor alleles of both genes bypass the spermiogenesis signal transduction pathway to which spe-27 belongs, resulting in premature spermiogenesis. In addition to spe-6 and spe-4, the inhibitory pathway involves multiple other genes: we are investigating 4 other suppressor mutations, at least two of which do not map to known spe genes (unpublished).
Our results suggest that SPE-4 affects multiple aspects of sperm development. Earlier results showed that it aids in the packaging of MSP in primary spermatocytes [22,23]. We show that SPE-4 is also involved in spermiogenesis, and that one of the functions is inhibitory, preventing premature spermiogenesis until the spermiogenesis activation signal is received by the spermatid. The defects we observed in spe-4(hc196) mutants include (i) accumulation of excess spermatocytes, (ii) premature activation of some spermatids, but (iii) an inability of other spermatids to activate, although many defective hermaphrodite produced self-spermatids can be transactivated male seminal fluid. Finally some male sperm display a fertilization defect (iv), even though they navigate successfully to the spermatheca. This constellation of defects is difficult to explain. It is as if the spe-4(hc196) phenotypic trajectory for spermatids is binary. Either the spermatids fail to activate, or they activate prematurely resulting in some normal sperm and others that cannot fertilize. The fact that many mutant spermatozoa are incapable of fertilization suggests that the timing of spermiogenesis is very important. Premature spermiogenesis may preclude proper segregation of cellular components required for fertilization.
The spe-4(hc196) mutation described here should allow us to gain a better understanding of the cellular function of this presenilin homolog. It retains the most function of any spe-4 allele to be studied, and its newly-discovered role inhibiting spermiogenesis was unexpected, given its MSP packaging function early in spermatogenesis. The inhibitory role spe-4 plays in spermiogenesis may involve known proteolytic and/or calcium regulatory aspects of presenilin function, or it may involve yet-to-be discovered presenilin functions.
Methods
Worm strains and culture
All C. elegans strains were maintained on Escherichia coli OP50-seeded Nematode Growth Media (NGM) agar plates and manipulated as described by Brenner [41]. The Caenorhabditis Genetic Center kindly provided the following strains: N2, CB4856, spe-27(it132ts)IV, spe-27(it132ts) unc-22(e66)IV, spe-29(it129) dpy-20(e1282ts)IV, him-5(e1490)V, spe-27(it132ts)IV; him-5(e1490)V, unc-22(e66)IV, dpy-5(e61)I; him-5(e1490)V, dpy-5(e61) unc-29(e193)I; spe-27(it132ts)IV, spe-8(hc53)I, spe-12(hc76)I, spe-19(hc41)V, fer-1(hc13ts)I, sDf5/spe-4(hc78) unc-15(e73)I. We received the strain bearing the hc196 mutation, which was isolated in a suppressor screen of spe-27(it132ts) [20,25], from Samuel Ward at the University of Arizona. We backcrossed hc196; spe-27(it132ts) unc-22(e66) six times with spe-27(it132ts), each time recovering unc-22 F2 that were fertile at 25°C. In addition to hc196, we also received numerous strains harboring mutations that suppress spe-27(it132ts) from Samuel Ward. The strains sDf5/spe-4(q347) unc-13(e1091)I and sDf5/dpy-5(e61) spe-4(hc81)I were kindly provided by Steven W. L'Hernault at Emory University.
Single-nucleotide-polymorphism-based mapping of hc196
We mapped hc196 using its spe-27(it132ts) suppression phenotype. We constructed a mapping strain which had spe-27(it132ts) backcrossed into the Hawaiian strain CB4856 six times (henceforth HA-spe-27). The mapping cross consisted of hc196; spe-27(it132ts) unc-22(e66) hermaphrodites mated to HA-spe-27 males. Virgin F1 progeny were allowed to self-fertilize at 15°C, where spe-27(it132ts) worms are fertile. We isolated 237 F2 worms (as L4 larvae) and allowed them to self-fertilize at 25°C, where only hc196 homozygotes are fertile in a spe-27 background. Forty-five of the F2 were identified as fertile. These fertiles were combined for bulk DNA extraction, as were another 45 sterile F2 worms chosen arbitrarily, following the protocol of Wicks et al. [24]. We initially mapped hc196 to chromosome I using SNP loci on the following clones (Chromosome: nucleotide position): pKP1119 (I: 8,508,270), T13C2 (II: 6,789,140), B0280 (III: 7,130,643), AC3 (V: 10,365,871). We did not analyze SNPs on chromosome IV because preliminary data indicated hc196 was unlinked to spe-27 on chromosome IV. The hc196 interval on Chromosome I was mapped using SNPs on the following clones (clone: genetic map location: nucleotide position): pkP1101 (C54G6: -17.40: 992,188), pkP1106 (Y54E10A: -4.47: 3,163,067), pkP1109 (E01A2: -1.66: 4,132,728), pkP1055 (C09D4: 0.05: 5,482,534), snp_F26B1[1] (F26B1: 0.98: 6,324,724), pkP1117 (T23G11: 2.26: 7,686,868), pkP1119 (K02B12: 2.88: 8,508,270), and pkP1071 (C37A5: 23.41: 14,154,876). All SNP information based upon WormBase release WS187.
Two- and three-factor mapping of hc196
To refine the SNP-mapped interval of hc196, we mapped the mutation in a trans-heterozygote cross with dpy-5. All strains had a spe-27 background, so that the hc196 phenotype was fertility. To further narrow the hc196 locus we performed a three-point mapping cross with dpy-5(e61)I (map locus 0.00) and unc-29(e193)I (map locus 3.32). These two genes were chosen because earlier map data suggested that they flank hc196. Male hc196; spe-27 males were mated to virgin dpy-5 unc-29; spe-27 hermaphrodites. Cross progeny (F1) were selfed at 15°C, and F2 recombinants (Dpy-non-Unc orUnc-non-Dpy) were isolated and raised at 15°C. F3 progeny from each recombinant F2 were checked for fertility at 25°C; those F3 that were fertile indicated that their F2 parent was heterozygous for hc196. The map locus was calculated based upon the number of recombination events between dpy-5 and hc196, and between hc196 and unc-29.
Complementation crosses
To identify additional alleles of the gene mutated in hc196, we tested a number of other uncharacterized spe-27 suppressor mutations. Because hc196 males have only weak fertility, we used heterozygous males (hc196/+; spe-27 unc-22/spe-27 +) and crossed them with hermaphrodites of the strain to be tested, such as hc197; spe-27 unc-22. We isolated virgin, non-Unc F1 at 25°C and assessed fertility. If the two mutations were allelic, half of the F1s should be sterile and half should be fertile (noncomplementation). Alternatively, if they were different genes, all F1s should be sterile (complementation). In addition to hc197, the other spe-27 suppressor alleles tested included hc166, hc176, hc199, hc201, hc204, hc205, zq6, zq7, and zq8. These suppressor strains were all in a spe-27 unc-22 background. We performed control complementation tests with three spe-6 alleles that are known to suppress spe-27 sterility (hc163, hc187, and hc188). Based on the outcome of the genetic mapping, we also performed a complementation test with spe-4, which maps to 2.10 on chromosome I, very near the hc196 locus. hc196; spe-27 males were mated to spe-4(hc78) unc-15 hermaphrodites at 20°C. Virgin F1 were isolated at 25°C to assess fertility. Control crosses with N2 males mated to spe-4 unc-15 hermaphrodites were also performed.
Molecular identification of hc196 as spe-4
We sequenced spe-4 from our hc196 worms using the Eppendorf TripleMaster PCR System™ for high fidelity PCR. The 2,852 bp fragment was amplified with primers F1 and R1 (Fig. 2) and extended from 451 bp upstream through 300 bp downstream of the spe-4 coding sequence. The PCR product was cloned using the AccepTor™ vector system and NovaBlue Singles™ competent cells (Novagen). Recombinant plasmids were purified from overnight cultures, and the insert was sequenced. In addition to the pETBlue up and pETBlue down vector primers, we constructed a set of sequencing primers dividing the insert into ~500 bp segments (Fig. 1). Automated DNA sequencing was performed by the Beckman Research Institute of the City of Hope (Duarte, CA) on a Hitachi AB model 3730 capillary DNA analyzer. The putative mutation identified by sequencing insert was verified by sequencing a PCR product amplified from hc196 genomic DNA containing the mutation.
Microinjection transformation rescue
We amplified the same 2.8 kb PCR product described above from wild-type worms. Wild-type worms were microinjected with the spe-4 PCR product (20 ng/μl) along with myo-2::GFP (100 ng/μl) as a transformation marker. Transformed worms were cultured and the progeny examined for transformation. We recovered a strain of worms that transmitted the transgenes stably across generations (zqEx1 [myo-2::GFP spe-4(+)]). Transformed males were mated to dpy-5 hc196; spe-27 unc-22 hermaphrodites. F1 worms with GFP expression were isolated and F2 worms that were GFP positive and dpy-5 but not unc-22 were isolated and kept at 25°C. Their fertility was scored daily. As a control, wild-type males were used instead of transformant males.
spe-4 suppression phenotype
We tested the ability of spe-4(hc196) to suppress spe-8(hc53), spe-19(hc41), and spe-29(it129), additional genes in the spermiogenesis activation pathway. To perform these tests, we constructed spe-4(hc196) double mutants with each of these genes and assessed their fertility at 25°C. We also tested the spe-27 suppression phenotype of three other spe-4 alleles that result in complete hermaphrodite sterility: hc78, hc81, and q347. Both hc81 and q347 are molecular null alleles, one being an amber stop and the other a nonsense deletion, respectively, while hc78 is a missense mutation that does not prevent production of SPE-4 protein [22]. For this assay, we crossed spe-27 unc-22/spe-27+ males with hermaphrodites of the three spe-4 lines. Each spe-4 line was marked with either an Unc or a Dpy morphological mutation (see Worm Strains section above for details). F1 hermaphrodites were selfed, and we recovered homozygous spe-27 F2 hermaphrodites (by picking unc-22 worms) of two classes : one class was homozygous for their particular spe-4 allele (homozygotes for the marker mutation), and the other class was not homozygous for the spe-4 allele (wild-type for the marker mutation). All F2 were reared at 25°C and assayed for fertility. Some of the F2 hermaphrodites that were not homozygous for spe-4 were fertile. In every case, they segregated either Unc or Dpy phenotypes in the F3, so they were spe-4 heterozygotes. Finally, dpy-5 spe-4(hc196)/+ +; spe-27 unc-22/spe-27 + males were mated into spe-4(hc78) unc-15(e73) hermaphrodites at 20°C. F1 were self-fertilized at 15°C and only unc-22 non-dpy-5 F2 were isolated at 25°C for fertility assay. These F2 were transheterozygotes for spe-4 (dpy-5 spe-4(hc196) +/+ spe-4(hc78) unc-15; spe-27 unc-22). The genotype was confirmed by observation of dpy-5 and unc-15 phenotypes in the F3.
Authors' contributions
RG conducted most of the experimentation and contributed to manuscript preparation. WSL participated in the single nucleotide polymorphism mapping and transformation rescue experiments. CWL contributed to the microscopy and manuscript preparation. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We are endebted to Paul Muhlrad, who improved the manuscript with his comments and discussions and who originally isolated the spe-4(hc196) mutation. We also thank Samuel Ward, who provided us with the large collection of spe-27 suppressor mutants from Paul's screen. We are grateful to Aidyl Gonzalez-Serricchio for technical assistance and Bernard Lakowski for the C. remanei SPE-4 sequence. Bernard Lakowski, Ángel A. Valdés, and Steven L'Hernault provided helpful discussions. Several worm strains were kindly provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources. This research was funded by a grant from the U.S. National Institutes of Health (Award 5 S06 GM053933-10 to CWL).
Contributor Information
Ryoko Gosney, Email: rmorikawa@csupomona.edu.
Wei-Siang Liau, Email: wliau@chem.ucla.edu.
Craig W LaMunyon, Email: cwlamunyon@csupomona.edu.
References
- Christen R, Schackmann RW, Shapiro BM. Elevation of the intracellular pH activates respiration and motility of sperm of the sea urchin, Strongylocentrotus purpuratus. J Biol Chem. 1982;257:14881–14890. [PubMed] [Google Scholar]
- Neill AT, Vacquier VD. Ligands and receptors mediating signal transduction in sea urchin spermatozoa. Reproduction. 2004;127:141–149. doi: 10.1530/rep.1.00085. [DOI] [PubMed] [Google Scholar]
- Schackmann RW, Shapiro BM. A partial sequence of ionic changes associated with the acrosome reaction of Strongylocentrotus purpuratus. Dev Biol. 1981;81:145–154. doi: 10.1016/0012-1606(81)90357-2. [DOI] [PubMed] [Google Scholar]
- Boatman DE, Robbins RS. Bicarbonate: carbon-dioxide regulation of sperm capacitation, hyperactivated motility, and acrosome reactions. Biol Reprod. 1991;44:806–813. doi: 10.1095/biolreprod44.5.806. [DOI] [PubMed] [Google Scholar]
- Gadella BM, Harrison RA. The capacitating agent bicarbonate induces protein kinase A-dependent changes in phospholipid transbilayer behavior in the sperm plasma membrane. Development. 2000;127:2407–2420. doi: 10.1242/dev.127.11.2407. [DOI] [PubMed] [Google Scholar]
- Gadella BM, Harrison RA. Capacitation induces cyclic adenosine 3',5'-monophosphate-dependent, but apoptosis-unrelated, exposure of aminophospholipids at the apical head plasma membrane of boar sperm cells. Biol Reprod. 2002;67:340–350. doi: 10.1095/biolreprod67.1.340. [DOI] [PubMed] [Google Scholar]
- Wennemuth G, Carlson AE, Harper AJ, Babcock DF. Bicarbonate actions on flagellar and Ca2+ -channel responses: initial events in sperm activation. Development. 2003;130:1317–1326. doi: 10.1242/dev.00353. [DOI] [PubMed] [Google Scholar]
- L'Hernault SW. Spermatogenesis. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editor. C elegans II. Cold Spring Harbor, NY USA , Cold Spring Harbor Laboratory Press; 1997. pp. 271–294. [PubMed] [Google Scholar]
- L'Hernault SW, Shakes DC, Ward S. Developmental genetics of chromosome I spermatogenesis-defective mutants in the nematode Caenorhabditis elegans. Genetics. 1988;120:435–452. doi: 10.1093/genetics/120.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield GM, Villeneuve AM. Regulation of sperm activation by SWM-1 is required for reproductive success of C. elegans males. Curr Biol. 2006;16:252–263. doi: 10.1016/j.cub.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Nelson GA, Ward S. Vesicle fusion, pseudopod extension and amoeboid motility are induced in nematode spermatids by the ionophore monensin. Cell. 1980;19:457–464. doi: 10.1016/0092-8674(80)90520-6. [DOI] [PubMed] [Google Scholar]
- Ward S, Hogan E, Nelson GA. The initiation of spermiogenesis in the nematode Caenorhabditis elegans. Dev Biol. 1983;98:70–79. doi: 10.1016/0012-1606(83)90336-6. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay J, Lee J, Lee J, Lee JI, Yu JR, Jee C, Cho JH, Jung S, Lee MH, Zannoni S, Singson A, Kim DH, Koo HS, Ahnn J. Calcineurin, a calcium/calmodulin-dependent protein phosphatase, is involved in movement, fertility, egg laying, and growth in Caenorhabditis elegans. Mol Biol Cell. 2002;13:3281–3293. doi: 10.1091/mbc.E02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington NL, Ward S. FER-1 regulates Ca2+ -mediated membrane fusion during C. elegans spermatogenesis. J Cell Sci. 2006;119:2552–2562. doi: 10.1242/jcs.02980. [DOI] [PubMed] [Google Scholar]
- Shakes DC, Ward S. Initiation of spermiogenesis in C. elegans: a pharmacological and genetic analysis. Dev Biol. 1989;134:189–200. doi: 10.1016/0012-1606(89)90088-2. [DOI] [PubMed] [Google Scholar]
- Nance J, Minniti AN, Sadler C, Ward S. spe-12 encodes a sperm cell surface protein that promotes spermiogenesis in Caenorhabditis elegans. Genetics. 1999;152:209–220. doi: 10.1093/genetics/152.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldziler B, Chatterjee I, Singson A. The genetic and molecular analysis of spe-19, a gene required for sperm activation in Caenorhabditis elegans. Dev Biol. 2005;283:424–436. doi: 10.1016/j.ydbio.2005.04.036. [DOI] [PubMed] [Google Scholar]
- Minniti AN, Sadler C, Ward S. Genetic and molecular analysis of spe-27, a gene required for spermiogenesis in Caenorhabditis elegans hermaphrodites. Genetics. 1996;143:213–223. doi: 10.1093/genetics/143.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance J, Davis EB, Ward S. spe-29 encodes a small predicted membrane protein required for the initiation of sperm activation in Caenorhabditis elegans. Genetics. 2000;156:1623–1633. doi: 10.1093/genetics/156.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad PJ, Ward S. Spermiogenesis Initiation in Caenorhabditis elegans Involves a Casein Kinase 1 Encoded by the spe-6 Gene. Genetics. 2002;161:143–155. doi: 10.1093/genetics/161.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkey JP, Jansma PL, Minniti AN, Ward S. The Caenorhabditis elegans spe-6 gene is required for major sperm protein assembly and shows second site non-complementation with an unlinked deficiency. Genetics. 1993;133:79–86. doi: 10.1093/genetics/133.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arduengo PM, Appleberry OK, Chuang P, L'Hernault SW. The presenilin protein family member SPE-4 localizes to an ER/Golgi derived organelle and is required for proper cytoplasmic partitioning during Caenorhabditis elegans spermatogenesis. J Cell Sci. 1998;111 :3645–3654. doi: 10.1242/jcs.111.24.3645. [DOI] [PubMed] [Google Scholar]
- L'Hernault SW, Arduengo PM. Mutation of a putative sperm membrane protein in Caenorhabditis elegans prevents sperm differentiation but not its associated meiotic divisions. J Cell Biol. 1992;119:55–68. doi: 10.1083/jcb.119.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- Muhlrad PJ. Molecular and Cellular Biology. Ph.D. PhD thesis. Tucson, AZ , University of Arizona; 2001. A Genetic and Molecular Analysis of Spermiogenesis Initiation in Caenorhabditis elegans; p. 192. [Google Scholar]
- Ward S, Miwa J. Characterization of a temperature-sensitive fertilization-defective mutant of the nematode Caenorhabditis elegans. Genetics. 1978;88:285–303. doi: 10.1093/genetics/88.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Eukaryotic Linear Motif resource for Functional Sites in Proteins http://elm.eu.org/
- De Strooper B. Loss-of-function presenilin mutations in Alzheimer disease. Talking Point on the role of presenilin mutations in Alzheimer disease. EMBO Rep. 2007;8:141–146. doi: 10.1038/sj.embor.7400897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdile G, Gandy SE, Martins RN. The role of presenilin and its interacting proteins in the biogenesis of Alzheimer's beta amyloid. Neurochem Res. 2007;32:609–623. doi: 10.1007/s11064-006-9131-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe MS. When loss is gain: reduced presenilin proteolytic function leads to increased Abeta42/Abeta40. Talking Point on the role of presenilin mutations in Alzheimer disease. EMBO Rep. 2007;8:136–140. doi: 10.1038/sj.embor.7400896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeter EH, Ilagan MX, Brunkan AL, Hecimovic S, Li YM, Xu M, Lewis HD, Saxena MT, De Strooper B, Coonrod A, Tomita T, Iwatsubo T, Moore CL, Goate A, Wolfe MS, Shearman M, Kopan R. A presenilin dimer at the core of the gamma-secretase enzyme: insights from parallel analysis of Notch 1 and APP proteolysis. Proc Natl Acad Sci U S A. 2003;100:13075–13080. doi: 10.1073/pnas.1735338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Kelleher RJ., 3rd The presenilin hypothesis of Alzheimer's disease: evidence for a loss-of-function pathogenic mechanism. Proc Natl Acad Sci U S A. 2007;104:403–409. doi: 10.1073/pnas.0608332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aturaliya RN, Fink JL, Davis MJ, Teasdale MS, Hanson KA, Miranda KC, Forrest AR, Grimmond SM, Suzuki H, Kanamori M, Kai C, Kawai J, Carninci P, Hayashizaki Y, Teasdale RD. Subcellular localization of mammalian type II membrane proteins. Traffic. 2006;7:613–625. doi: 10.1111/j.1600-0854.2006.00407.x. [DOI] [PubMed] [Google Scholar]
- Cowburn RF, Popescu BO, Ankarcrona M, Dehvari N, Cedazo-Minguez A. Presenilin-mediated signal transduction. Physiol Behav. 2007;92:93–97. doi: 10.1016/j.physbeh.2007.05.053. [DOI] [PubMed] [Google Scholar]
- Guo Q, Furukawa K, Sopher BL, Pham DG, Xie J, Robinson N, Martin GM, Mattson MP. Alzheimer's PS-1 mutation perturbs calcium homeostasis and sensitizes PC12 cells to death induced by amyloid beta-peptide. Neuroreport. 1996;8:379–383. doi: 10.1097/00001756-199612200-00074. [DOI] [PubMed] [Google Scholar]
- Nelson O, Tu H, Lei T, Bentahir M, de Strooper B, Bezprozvanny I. Familial Alzheimer disease-linked mutations specifically disrupt Ca2+ leak function of presenilin 1. J Clin Invest. 2007;117:1230–1239. doi: 10.1172/JCI30447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landman N, Jeong SY, Shin SY, Voronov SV, Serban G, Kang MS, Park MK, Di Paolo G, Chung S, Kim TW. Presenilin mutations linked to familial Alzheimer's disease cause an imbalance in phosphatidylinositol 4,5-bisphosphate metabolism. Proc Natl Acad Sci U S A. 2006;103:19524–19529. doi: 10.1073/pnas.0604954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh BC, Hille B. Regulation of ion channels by phosphatidylinositol 4,5-bisphosphate. Curr Opin Neurobiol. 2005;15:370–378. doi: 10.1016/j.conb.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Tu H, Nelson O, Bezprozvanny A, Wang Z, Lee SF, Hao YH, Serneels L, De Strooper B, Yu G, Bezprozvanny I. Presenilins form ER Ca2+ leak channels, a function disrupted by familial Alzheimer's disease-linked mutations. Cell. 2006;126:981–993. doi: 10.1016/j.cell.2006.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XZ, Sternberg PW. A C. elegans sperm TRP protein required for sperm-egg interactions during fertilization. Cell. 2003;114:285–297. doi: 10.1016/S0092-8674(03)00565-8. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TMpred - Prediction of Transmembrane Regions and Orientation http://www.ch.embnet.org/software/TMPRED_form.html
- Liau WS, Gonzalez-Serricchio A, Deshommes C, Chin K, LaMunyon CW. A persistent mitochondrial deletion reduces fitness and sperm performance in heteroplasmic populations of C. elegans. BMC Genet. 2007;8:8. doi: 10.1186/1471-2156-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]