Abstract
Starvation of yeast cultures limited by auxotrophic requirements results in glucose wasting and failure to achieve prompt cell-cycle arrest when compared with starvation for basic natural nutrients like phosphate or sulfate. We measured the survival of yeast auxotrophs upon starvation for different nutrients and found substantial differences: When deprived of leucine or uracil, viability declined exponentially with a half-life of <2 days, whereas when the same strains were deprived of phosphate or sulfate, the half-life was ≈10 days. The survival rates of nongrowing auxotrophs deprived of uracil or leucine depended on the carbon source available during starvation, but were independent of the carbon source during prior growth. We performed an enrichment procedure for mutants that suppress lethality during auxotrophy starvation. We repeatedly found loss-of-function mutations in a number of functionally related genes. Mutations in PPM1, which methylates protein phosphatase 2A, and target of rapamycin (TOR1) were characterized further. Deletion of PPM1 almost completely suppressed the rapid lethality and substantially suppressed glucose wasting during starvation for leucine or uracil. Suppression by a deletion of TOR1 was less complete. We suggest that, similar to the Warburg effect observed in tumor cells, starving yeast auxotrophs wastes glucose as a consequence of the failure of conserved growth control pathways. Furthermore, we suggest that our results on condition-dependent chronological lifespan have important implications for the interpretation and design of studies on chronological aging.
Keywords: auxotrophy starvation, chronological aging, Warburg effect
To grow and survive, all cells must sense and respond to the availability of nutrients in their environment. In budding yeast (Saccharomyces cerevisiae), the means by which cells sense and respond to the availability of sources of nitrogen, phosphate, sulfate, and fermentable sugars are well studied. The nutrient sensor and assimilation pathways are known to dynamically modify activities of diverse cellular functions, including nutrient utilization, ribosome biosynthesis, and stress responses (1–4). The onset of cell division is directly dependent on the output of these pathways: When inadequate carbon, nitrogen, sulfur, or phosphorus is detected in its environment, it arrests at the start (G0/G1) of the cell cycle (5–7). Only under favorable conditions will progression through the cell division cycle proceed (8).
The failure to commit to the cell division cycle, and entering instead a resting or quiescent state, has always been assumed to be a survival strategy in the face of inadequate nutrition (6). This assumption is entirely consistent with findings that pathways that stimulate growth and division in nutrient-rich conditions, like target of rapamycin (TOR) and protein kinase A (PKA), repress entry into a resting state. Moreover, pathways that activate entry into a resting state, such as protein kinase C (PKC) and Snf1, are repressed by glucose, arguably the strongest activator of growth (4, 9). Thus, it came as no surprise that mutations in genes involved in these signaling pathways and their downstream targets have been found to affect long-term viability (9–12).
In a comparison with several nutrients, Saldanha et al. (13) observed that cells respond differently to starvation for a nutrient made necessary by mutation (leucine in a leucine auxotroph or uracil in a uracil auxotroph), compared with a “natural” starvation for phosphate or sulfate. Notably, in the latter cases, nearly all of the cells in the culture appeared to have arrested in G0/G1 (unbudded), whereas there was no similarly rapid, coordinated arrest upon starvation for leucine and uracil in the respective auxotrophs. In contrast, when cultures of exponentially growing cells in nutrient-limited chemostats were compared with the same limiting nutrients (2), no difference in the budding index between the natural and auxotrophic limitations was observed. Brauer et al. (2), however, observed that chemostat cultures limited for uracil or leucine differed from those limited by phosphate, sulfate, or ammonium in another way: The latter conserved much of the excess glucose available to them, whereas the former completely fermented the excess glucose to ethanol.
Both the lack of a uniform arrest during starvation of auxotrophs and the wasting of excess glucose suggest a failure of regulation. Specifically, they raise the possibility that one or more signals given during starvation for the natural nutrients, leading to cell-cycle arrest and cessation of glucose fermentation, are lacking when a nutrient made necessary only by a mutation runs out. It seemed likely that this failure of regulation might have consequences for the viability of cells that were starved of auxotrophic nutrients.
Here we show that starvation does indeed have differential effects on viability depending on the nature of the missing nutrient. Several auxotrophic strains we tested die exponentially when deprived of their cognate nutrients, whereas the same strains survive when deprived of phosphate or sulfate; the degree of the difference depends on the carbon source. We selected mutants that survive auxotrophic starvation for leucine and repeatedly found null mutations in PPM1 and TOR1. Our data further point to a major involvement of protein phosphatase 2A (PPA2) as a central component in the response upon auxotrophy starvation.
Results
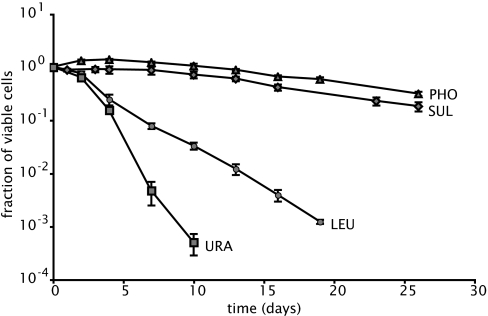
We determined the viability of yeast cells in dilute, nonsaturating media, where only a single nutrient limits growth. The survival of cells starved for phosphate, sulfate, uracil, or leucine was tested in triplicate in a single strain (DBY8863) doubly auxotrophic for leucine and uracil. In each case, this strain was growing in the appropriate nutrient-limited medium before being completely deprived of the nutrient by dilution, as described in Materials and Methods. Fig. 1 shows that the cells survive for weeks when starved of phosphate or sulfate (viability declines exponentially with a half-life of ≈10 days), whereas the cells starved of uracil or leucine die exponentially at a much higher rate (viability declines with a half-life of 0.8 or 1.9 days, respectively).
Fig. 1.
Survival of strain DB8863, measured as colony-forming units, during starvation for phosphate (triangles), sulfate (diamonds), leucine (circles), and uracil (squares). The number of viable cells was normalized to 100% and measured at the onset of starvation (day 0). The average values and SD for three independent replicate experiments are shown. Survival of prototrophs in limiting phosphate or sulfate is identical to that of auxotrophs and is unaffected by added uracil or leucine (data not shown).
This limitation-dependent difference in survival extends previous observations of other significant differences in the physiological responses to starvation for sulfate or phosphate, on the one hand, and to leucine or uracil, on the other hand. Brauer et al. (2) found that leucine or uracil limitation in chemostat cultures results in an excess glucose consumption, compared with phosphate or sulfate limitation. Saldanha et al. (13) found a relative failure to arrest the cell cycle promptly during leucine or uracil starvation, compared with the prompt arrest at G0/G1 that is found during phosphate or sulfate starvation. Supporting information (SI) Fig. S1 shows two typical experiments showing the excess consumption of glucose by starving auxotrophs. These phenomena do not appear to depend on the yeast strain background: We have observed them in S288c-, CEN.PK-, and W303-derived strains.
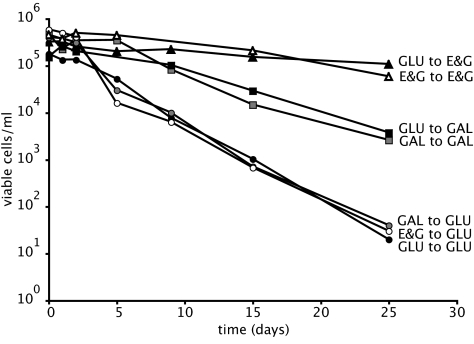
Because of the potential connection between survival and glucose consumption, we tested survival during leucine starvation in media containing either galactose or a combination of ethanol and glycerol instead of glucose as carbon sources. Fig. 2 shows that the nature of the carbon source has a significant impact on the rate of loss of viability during leucine starvation. In galactose, the viability declines (half-life of 2.8 days) significantly more slowly than in glucose (half-life of 1.9 days). In ethanol/glycerol, viability during leucine starvation is even higher and declines at about the same rate as that found for phosphate starvation in glucose (11 days).
Fig. 2.
Survival of strain DB8863, measured as colony-forming units, during leucine starvation in media containing as carbon source ethanol and glycerol (triangles), galactose (squares), or glucose (circles). Cells were pregrown on leucine-limited media containing, as a carbon source, glucose (black symbols), galactose (gray), or ethanol and glycerol (white).
A series of experiments determined that these differences depend only on the carbon source in which the cells are starving not on their prior growth conditions. When cells were pregrown on leucine-limited glucose media and transferred to leucine-limited media with galactose, they displayed the same death rates as when the cells were both growing and starving in galactose medium. The same holds true for the transfer to ethanol/glycerol media and the reciprocal switch: Death rates in leucine-limited glucose medium were independent of the carbon source during prior growth.
To investigate the mechanisms of sensitivity to auxotrophy starvation, we performed a genetic screen for mutants with enhanced survival upon leucine starvation (see Materials and Methods). Briefly, this process involved allowing independent cultures to starve for leucine until viability declined ≈200,000-fold, regrowing cultures in limiting phosphate and excess leucine, and repeating the starvation for leucine. After five to seven rounds of starvation and recovery, single colonies were isolated and tested for viability in media lacking leucine. These viability assays were performed, as in Fig. 1, at low cell density (≈3 × 105 per ml) to avoid acidification of the medium or depletion of nutrients other than leucine.
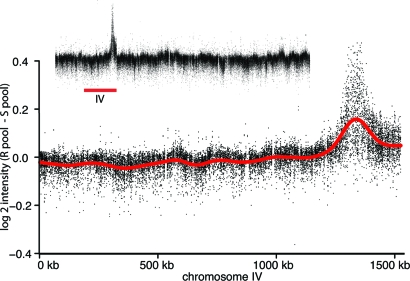
Initially, two independent mutants with enhanced viability during leucine starvation were isolated (strains DBY11202 and DBY11203). Complementation tests revealed that these mutants do not complement each other, implicating a single locus. One of the mutants, DBY11202, was used for both array-assisted bulk segregant analysis (14) and mutation detection by using overlapping tiling arrays of the entire yeast genome (15). The bulk segregant analysis identified one linkage peak on chromosome IV (Fig. 3). Within that region, tiling arrays analysis predicted one polymorphism within the gene PPM1 (Table 1). The PPM1 gene was sequenced in DBY11202 and DBY11203, and in both strains, we found frameshift mutations introducing premature STOP codons (a single-bp deletion at bp 1,333,843 introducing a STOP codon at L49 in DBY11202 and a 71-bp deletion starting at bp 1,333,461 introducing a STOP codon at L146 in DBY11203).
Fig. 3.
Bulk segregant analysis on tiling arrays reveals a single locus on chromosome IV. The difference in signal intensity (resistant pool–sensitive pool) is shown for chromosome IV and the whole genome (Inset).
Table 1.
Viability during nutrient starvation of wild-type and mutant strains
| Strain | Limitation | Carbon source | Half-life, days |
|---|---|---|---|
| DBY8863 | Phosphate | Glucose | 11 |
| Sulfate | Glucose | 9.4 | |
| Leucine | Glucose | 1.9 | |
| Uracil | Glucose | 0.8 | |
| Leucine | Galactose | 2.8 | |
| Leucine | Ethanol plus glycerol | 11 | |
| BY4742 | Leucine | Glucose | 2.0 |
| Uracil | Glucose | 0.7 | |
| ppm1Δ0 | Leucine | Glucose | 8.6 |
| Uracil | Glucose | 2.6 | |
| tor1Δ0 | Leucine | Glucose | 2.4 |
| Uracil | Glucose | 1.0 |
Another 12 independent mutants were isolated, and all were resequenced by using tiling arrays and Sanger sequencing (15). The mutations that were found in annotated genes are listed in Table 2. Mutations in some genes were found more than once and thus are likely to be causative. In addition to the two truncating ppm1 mutations, we found three independent truncating mutations in TOR1, one truncating and one missense mutation in SCH9, one nonsense and one missense mutation in HRD1, and one nonsense and one missense mutation in HAP1. Initial tests of the ability of these spontaneous mutant strains to survive leucine starvation suggested that the ppm1 mutations had the strongest effect of restoring survival to levels found with sulfate or phosphate starvation, followed by sch9 and tor1 mutations; the hap1 and hrd1 mutations had very minor effects. Below, we further characterize the effect of loss of function of ppm1 and tor1, recognizing that it is likely that many of the other genes in which we recovered mutations also may be involved in starvation survival. Specifically, the sch9 mutants seem to confer significant resistance (half-life on leucine-limited glucose medium is >5 days); however, the severe growth defect of the sch9 deletion strain (16) compromises a straightforward comparison.
Table 2.
Mutations found in isolated strains with enhanced survival upon leucine starvation
| Gene | Mutation (chr:bp) | Protein | Parent | Strain | Other mutations in |
|---|---|---|---|---|---|
| PPM1 | 4:1333843 A to Δ | L49X | 1 | DBY11202 | CPR3, GCD1, ACF4, KEX1 |
| PPM1 | 4:1333461 71bpΔ | L146X | 1 | DBY11203 | |
| TOR1 | 10:559755 C to Δ | S116X | 1 | DBY11204 | |
| TOR1 | 10:563920* | S1509X | 1 | DBY11205 | |
| TOR1 | 10:565483 C to G | Y2025X | 1 | DBY11206 | ADP1 |
| SCH9 | 8:511331 G to A | G657S | 2 | DBY11211 | SSK22 |
| SCH9 | 8:509689 C to AA | N125X | 2 | DBY11214 | |
| HRD1 | 15:302637 C to T | Q134X | 1 | DBY11207 | |
| HRD1 | 15:301850 G to T | C396F | 1 | DBY11208 | CDC60 |
| HAP1 | 12:646559 C to G | S48X | 2 | DBY11213 | AUS1 |
| HAP1 | 12:647879 T to G | F488C | 2 | DBY11215 | |
| KOG1 | 8:480255 T to C | S140 P | 2 | DBY11212 | |
| TPD3 | 1:126722 G to C | D615H | 1 | DBY11210 | FLO1, FLO5 |
| HRD3 | 12:557609 G to T | G274C | 1 | DBY11210 | FLO1, FLO5 |
| 1 | DBY11209 | UBR2, 15:1042k to end |
Genes contributing to the gene neighborhood (Fig. 6) are given in the first column. Parent 1, FY strain DBY8863, parent 2, SK1 strain NKY1060. These 14 strains were randomly chosen from 27 independent selections.
*TTACGA to CTTCTTA
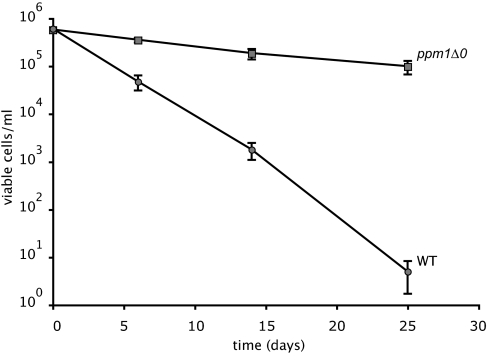
To confirm more rigorously that the ppm1 and tor1 mutations and not other bp differences (whether detected or not) in the strains we recovered are responsible for the enhanced survival, we tested the survival of constructed ppm1- and tor1-deletion mutants. Fig. 4 shows survival during leucine starvation in a ppm1-deletion mutant and the isogenic parent DBY8863. Fig. S2 shows results for a ppm1- and tor1-deletion strain from the systematic yeast-deletion project (17), tested for survival upon leucine and uracil starvation; the half-lives also are given in Table 1. The isogenic parent strain (BY4742) dies at about the same rate as the closely related strain DBY8663, which was the parental strain in our selection (2.0 days for leucine and 0.7 day for uracil). Survival of the ppm1Δ0 mutant is considerably higher than wild type (half-life of 8.6 days for leucine and 2.6 days for uracil). Survival of the tor1Δ0 mutant is only slightly higher than wild type (half-life of 2.4 days for leucine and 1 day for uracil). These results strongly support a causal role for ppm1-null mutations in survival upon auxotrophy starvation, which is consistent with the repeated recovery of null mutations in the selection. The situation for tor1 mutants is less clear. The three independent spontaneous tor1 mutants we recovered had half-lives of 3.9 (±0.3) days on leucine-limited media. The modest suppression of the tor1 deletion in the BY background could be due to strain differences (e.g., because of the relative contribution of Tor2p in TOR complex 1) or, alternatively, mutations that were not detected in our screen.
Fig. 4.
Survival of strain DBY8863 (circles) and isogenic ppm1Δ0 derivative (squares) during leucine starvation, with mean and SD of six independent replicates.
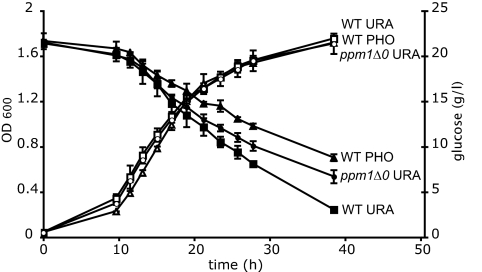
Finally, we tested whether the ppm1 mutations had any effect on the glucose-wasting phenomenon previously associated with starvation for leucine and uracil, compared with starvation for phosphate or sulfate. Fig. 5 shows that, during uracil starvation, our ppm1-deletion strain consumes significantly less excess glucose than its isogenic parent, but more compared with exponential growth of the parent strain during phosphate starvation. Fig. S3 shows the same for leucine starvation.
Fig. 5.
Growth and glucose consumption during phosphate and uracil starvation. Triangles, strain DBY8863 during phosphate starvation; squares, strain DBY8863 during uracil starvation; circles, isogenic ppm1Δ0 deletion strain during uracil starvation; open symbols, OD600; closed symbols, glucose concentration. Values represent averages and SD of two (DBY8863) or four (ppm1Δ0 deletion strain) independent experiments.
Discussion
Survival of Nongrowing Cells Depends on the Nutritional Limitation and the Carbon Source.
Our results show that starvation for auxotrophic requirements is substantially more lethal than starvation for phosphate or sulfate. Just by changing the limiting nutrient, we found ≥10-fold differences in exponential survival rate (e.g., between phosphate and uracil starvation in Fig. 1). Hence, an important aspect of chronological aging has gone largely unnoticed so far: Yeast viability depends on which nutrient is limited first. Therefore, it seems clear that rigorous control of conditions is required while studying chronological aging specifically because gene effects previously found to affect chronological aging (10, 12) are often smaller than the conditional effects we have found here. Moreover, all studies on chronological aging in S. cerevisiae published so far have made use of auxotrophic strains (usually including leucine, histidine, and uracil as auxotrophic requirements). Assays that have been used in determining chronological lifespan almost uniformly involved growing auxotrophic strains to high density in YPD or synthetic complete medium and subsequent starvation in the same medium. Alternatively, cells were transferred to water after growth to saturation (reviewed in refs. 18 and 19). In both experimental designs, one cannot tell which nutrient runs out first, and it is unclear to the absence of what nutrient(s) the cells responds. As we have shown, the nature of the limiting nutrient determines lifespan. Thus, the techniques of measurement of chronological aging require a rigorous reassessment.
The rapid loss of viability during starvation for leucine is, in part, dependent on the carbon source(s) available during starvation. Changing the carbon source from glucose to galactose or ethanol/glycerol, which are less preferred sources of carbon, drastically increased the lifespan during starvation. These results strengthen the notion that growth-stimulating pathways that are activated by glucose (TOR and PKA) have a negative effect on lifespan (9, 12, 20), whereas glucose represses pathways that activate entry into a resting state (4, 9). It is significant that the carbon source matters only during the starvation: The carbon source available before starvation makes no detectable difference, ruling out prior metabolic activity or growth rate as a factor influencing viability.
Amino Acid Starvation and the Cell Division Cycle.
Survival during starvation of auxotrophs has been studied previously in a number of organisms, including Neurospora (21), Escherichia coli (22), and S. cerevisiae (23, 24), with varying survival rates. In S. cerevisiae, Henry (24) reported very rapid loss of viability of S. cerevisiae inositol auxotrophs (hundreds of fold per day) and only a modest loss of viability upon lysine, adenine and tryptophan starvation. The rates of loss of viability found by Henry (24) for amino acid starvation appear to be comparable to our observation for leucine and uracil; for inositol starvation, the rate is markedly higher. We also tested histidine auxotrophs with very similar results (data not shown). An exception to the pattern that yeast auxotrophs die rapidly upon starvation seems to be methionine. When Unger and Hartwell (5) observed that methionine starvation of a methionine auxotroph results in G0/G1 arrest, they reported no loss of viability. They proposed that methionine or a product of methionine (e.g., methionyl-tRNA) acts as a more general signal for starvation and also is involved in signaling sulfate starvation (5).
Following the reasoning of Unger and Hartwell (5), the lethality observed with auxotrophic starvations results, in part, from a failure to execute a cell-cycle arrest; the prompt arrest of the cell cycle during starvation for methionine or the natural nutrients phosphate or sulfate serves to put cells into a “safe” quiescent state. This idea is in accord with our observation that auxotrophs starved for phosphate or sulfate results in low lethality. We suppose further that the lethality depends on metabolic activities (e.g., initiation of DNA replication) inappropriate to the circumstances that happen to a greater degree when good carbon sources are available.
Loss-of-Function Mutations in a Number of Genes Can Suppress Lethality During Auxotrophy Starvation.
Identifying the genetic basis of new phenotypes is a serious challenge in biological studies. With the use of recently developed microarray technology, we were able to identify causal mutations in strains with acquired longer lifespans. The mutations that we recovered that suppress the lethality of auxotrophic starvation provide clues to the identity of regulatory pathways involved. We repeatedly isolated mutations in three genes (PPM1, TOR1, and SCH9) that significantly suppress the lethality phenotype. Two independently derived ppm1 deletions suppress lethality, resulting in survival over time close to that observed for phosphate starvation. The ppm1 deletion also suppresses the elevated glucose consumption during auxotrophy-limited growth, although not completely. In the case of TOR1, a deletion suppresses the lethality phenotype, but significantly less well.
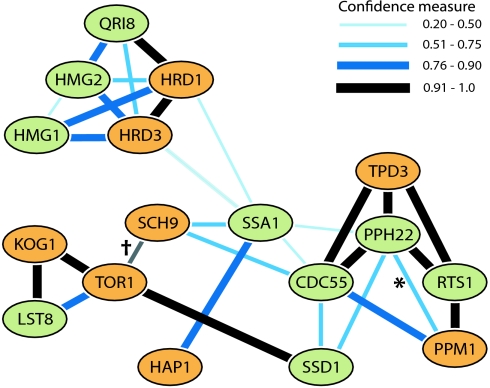
In addition to the use of new microarray technology, we also made use of recently developed bioinformatic tools. The bioPIXIE program (25) summarizes evidence for gene interactions by using Bayesian analysis of diverse kinds of experimental data, including coexpression, synthetic lethality, and more direct biochemical protein interaction data. Fig. 6 summarizes the genes in which we found mutations and the closest associated genes, as inferred by using the bioPIXIE program. We found that, in many mutants, the suppression of lethality is the result of a loss of function of one of a group of genes that have already been implicated in signaling pathways that regulate one or more basic cellular processes: metabolism, the cell cycle, growth rate, and stress response (26–28).
Fig. 6.
Gene neighborhood of genes affected by mutation in the leucine survival screen (orange) and the top-ranking associated genes (green). *, Methylation site; †, added interaction based on evidence published previously (39). See Materials and Methods for details.
PP2A Plays an Important Role in Survival upon Auxotrophy Starvation.
The PPM1 gene encodes a carboxyl methyl transferase that methylates the catalytic subunit of PP2A (in yeast, encoded by PPH21 or PPH22). This function is the only known type of this protein, and Ppm1p appears to be the only protein in yeast that carries out this methylation (29, 30). Ppm1p and its substrate, the C-terminal domain of the catalytic subunit, are both highly conserved in eukaryotes from yeast to human. Loss of function of Ppm1p in yeast greatly reduces the interaction of the catalytic and regulatory subunits of PP2A (29, 30) and results in a rapamycin-resistant phenotype (30). The administration of rapamycin mimics nutrient-poor conditions; loss of function of the TOR complex1 has a similar effect. As is clear from Fig. 6, the set of genes we recovered are all closely related to each other, and many of them could, like ppm1 and tor1 mutations, mimic the perception of a nutrient-poor environment and set off a starvation response, thus leading to an increase in long-term viability and a decrease in glucose consumption.
We found that, contrary to the wild type, survival upon auxotrophy starvation is independent of rapamycin in the ppm1Δ0 strain (Fig. S4). The rapamycin-resistant phenotype of a ppm1Δ0 mutant has been observed previously, and it is likely mediated through Tap42p, which binds both TOR complex1 and the unbound PP2A catalytic subunits Pph21p and Pph22p (30, 31). Inactivation of TOR complex1 leads to the dissociation of the PP2A complex from Tap42p (31–33), whereas inactivation of Ppm1p leads to the dissociation of the PP2A catalytic subunits from the PP2A regulatory and scaffolding subunits (29, 30, 34). The mutation we found in TPD3 also might lead to reduced stability of the PP2A trimeric complex. Hence, we can speculate that ultimately it is elevated levels of unbound PP2A catalytic subunit that result in an auxotrophy starvation-resistant phenotype. However, how TOR complex1, Sch9p and PP2A, or unbound Pph21p or Pph22p act in the sensitivity to auxotrophy starvation is not yet clear.
Starving Yeast Auxotrophs and Human Tumor Cells May Waste Glucose (Warburg Effect) Through the Failure of Conserved Growth Control Pathways.
An interesting comparison can be made between the results we have found in yeast and the properties of human tumor cells. An inference arises from the coincidence of three factors: cell-cycle dysregulation [found in both yeast auxotrophs (2, 13) and in tumor cells], the genes involved (conserved growth regulators in both yeast and animal cells and implicated in tumor etiology), and the relatively new observation of excess glucose consumption (which we call “wasting”) known as the Warburg effect (35). This elevated glucose consumption is routinely used to identify tumors in vivo with the use of the glucose analogue 2-[18F]fluoro-2-deoxy-d-glucose. It seems that both tumor and yeast cells growing under auxotrophy starvation grow in an uncontrolled state of increased metabolism without the proper checks and balances. In both yeast and humans, TOR1 could be an important effector of this phenotype because mammalian TOR1 (mTOR1) is a protooncogene, and constitutively active mTOR complex1 leads to tumor growth. We found that loss of function of yeast TOR1 leads to increased survival upon auxotrophy starvation, implicating active Tor1 in the sensitivity toward auxotrophy starvation. Inhibiting TOR complex1 activity with rapamycin increases survival upon auxotrophy starvation (Fig. S4). The same analogy can be drawn for mammalian kinases Akt1p and S6K1, also protooncogenes (36, 37), and their yeast homologue Sch9p, which we recovered twice in our screen. In light of this and given the highly evolutionary conserved C-terminal domain of protein phosphatase 2A, it would be of great interest to see how the protein phosphatase methyltransferase affects cell signaling, in both yeast and human.
To conclude, our results show that the survival of yeast cells that have stopped growing for nutritional reasons depends on why they ceased growing. If natural nutrients, like phosphate or sulfate, are depleted, a prompt and orderly arrest of the cell division cycle, accompanied by a cessation of glucose fermentation, ensures that cells are able to survive for weeks. Depletion of nutrients made necessary because of an auxotrophic mutation, like leucine or uracil (but probably not methionine), does not lead to an orderly arrest. Glucose consumption remains high even after cessation of growth, and the cells die exponentially in a matter of days. This rapid lethality can be suppressed by the loss of a number of proteins already implicated in sensation of nutrient sufficiency and/or growth control, such as the products of the PPM1, SCH9, and TOR1 genes.
Materials and Methods
Yeast Strains and Growth Conditions.
FY derivative strain DBY8863 (MATa leu2-Δ1 ura3-52) and SK1 derivative strain NKY1060 (MATalpha ho::LYS2 lys2 ura3 leu2::hisG) were used for the selection of mutants, linkage analysis, and growth experiments. DBY8863 and BY4742 (MATalpha his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) and their single-deletion derivatives for ppm1Δ0 and tor1Δ0 (BY4742 only) were used for the confirmation of survival phenotypes.
All minimal media were slightly adapted from Saldanha et al. (13). Deviations were as follows: for phosphate limitation, 1 g/liter KCl and 10 mg/liter KH2PO4; for sulfate limitation, 2.6 mg/liter (NH4)2SO4; for uracil limitation, 4 mg/liter uracil; and for leucine limitation, 40 mg/liter leucine. Where needed, excess additions of 240 mg/liter leucine, 40 mg/liter uracil, 40 mg/liter histidine, and 60 mg/liter lysine were used. All media contained 20 g/liter glucose or, when indicated, 20 g/liter galactose or 8.1 g/liter ethanol and 12.6 g/liter glycerol as a carbon source. YPD contained 10 g/liter bacto yeast extract, 20 g/liter bacto peptone, and 20 g/liter glucose.
For growth experiments, cultures were grown at 30°C, agitated at 200 rpm, in 250-ml erlenmeyer flasks with a working volume of 50 ml. Cultures were pregrown overnight on the same media and diluted at t = 0 to an OD600 of 0.04. For glucose determinations, 1 ml of culture was centrifuged at 13,000 × g, and supernatants were stored at −20°C. Glucose was determined enzymatically (in triplicate) by using a d-Glucose commercial kit (R-Biopharm).
Survival Assays.
A single colony was inoculated and grown in 3 ml of YPD. After 24 h, this culture was diluted 50-fold in nutrient-limited medium. After 24 h, this culture was diluted 150-fold in media lacking entirely the appropriate nutrient (time 0), and samples were plated on YPD agar plates to determine colony-forming units. In sulfate-limited conditions, there was some growth after the last transfer due to the presence of trace amounts of copper, manganese, and zinc sulfate, and, hence, time 0 was set 2 days after the last transfer.
Enrichment for Mutants with Enhanced Survival upon Leucine Starvation.
Strains DBY8863 and NKY1060 were grown in leucine-limited medium, and viability was assayed over time. When the total fraction of viable cells was <5 × 10−4, ≈ 100–1,000 viable cells were transferred to phosphate-limited medium to limit selection for strains that simply grow faster or recover from arrest more quickly in the selection media. After several days, this culture was used as an inocculum for a new round of leucine starvation. Between five and seven rounds of enrichment were performed, after which single colonies were isolated and assayed for viability.
Mutation Detection.
DNA of one clone of each enrichment was hybridized to Affymetrix yeast tiling arrays and analyzed for mutations as described previously (15). SNP predictions in ORFs were confirmed by PCR and Sanger sequencing (in duplicate).
Linkage Analysis.
Linkage analysis was performed by using a strategy described previously (14). In short, strain DBY11202 was backcrossed twice to get a 2:2 segregation (resistant; high viability upon leucine starvation, sensitive; low viability). One of the resistant segregants was crossed to the sensitive, divergent SK1 derivative strain NKY1060. After sporulation, segregants were scored for viability upon leucine starvation. DNA was isolated from the resistant and sensitive pools (segregants with intermediate resistance were omitted) and hybridized to Affymetrix tiling arrays (15). For estimation of the linkage peak, only the probes were used that have known differences between FY and SK1 strains (38).
Assembly of the Gene Neighborhood (Fig. 6).
To place the mutations found in the genetic screen into a biological context, we used a program called bioPIXIE (25). All genes affected by mutations (Table 2, excluding CPR3, GCD1, ACF4, and KEX1 because in this strain it was established that PPM1 is causal) were used as input in bioPIXIE to assemble the gene neighborhood and to generate the accompanying gene rank list (Table S1). The top nine ranking genes of the gene neighborhood remained identical when only using a subset of all genes (Table 2, column 1) as input. Hence, this subset contributes most strongly to the gene neighborhood. The graph obtained from bioPIXIE was redrawn retaining the gene interaction information between the top-ranking genes in the neighborhood and the genes in Table 2. The connection between TOR1 and SCH9 was added based on evidence published previously (39).
Supplementary Material
Acknowledgments.
We thank D. Gresham, D. Ruderfer, J. Schacherer, and J. Shapiro for help and advice on tiling array analysis; R. Koszul (Harvard University, Cambridge, MA) for providing strain NKY1060; and various colleagues for critical reading of the manuscript. This work was supported by the National Institute of General Medical Sciences Center for Quantitative Biology/National Institutes of Health Grant P50 GM-071508 and National Institutes of Health Grant R01 GM-046406 (to D.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802601105/DCSupplemental.
References
- 1.Boer VM, de Winde JH, Pronk JT, Piper MD. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J Biol Chem. 2003;278:3265–3274. doi: 10.1074/jbc.M209759200. [DOI] [PubMed] [Google Scholar]
- 2.Brauer MJ, et al. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- 4.Schneper L, Duvel K, Broach JR. Sense and sensibility: Nutritional response and signal integration in yeast. Curr Opin Microbiol. 2004;7:624–630. doi: 10.1016/j.mib.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Unger MW, Hartwell LH. Control of cell division in Saccharomyces cerevisiae by methionyl-tRNA. Proc Natl Acad Sci USA. 1976;73:1664–1668. doi: 10.1073/pnas.73.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Werner-Washburne M, Braun E, Johnston GC, Singer RA. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol Rev. 1993;57:383–401. doi: 10.1128/mr.57.2.383-401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wanke V, Pedruzzi I, Cameroni E, Dubouloz F, De Virgilio C. Regulation of G0 entry by the Pho80-Pho85 cyclin-CDK complex. EMBO J. 2005;24:4271–4278. doi: 10.1038/sj.emboj.7600889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jorgensen P, Tyers M. How cells coordinate growth and division. Curr Biol. 2004;14:R1014–R1027. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 9.Gray JV, et al. “Sleeping beauty”: Quiescence in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2004;68:187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- 11.Krause SA, Gray JV. The protein kinase C pathway is required for viability in quiescence in Saccharomyces cerevisiae. Curr Biol. 2002;12:588–593. doi: 10.1016/s0960-9822(02)00760-1. [DOI] [PubMed] [Google Scholar]
- 12.Powers RW, III, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saldanha AJ, Brauer MJ, Botstein D. Nutritional homeostasis in batch and steady-state culture of yeast. Mol Biol Cell. 2004;15:4089–4104. doi: 10.1091/mbc.E04-04-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brauer MJ, Christianson CM, Pai DA, Dunham MJ. Mapping novel traits by array-assisted bulk segregant analysis in Saccharomyces cerevisiae. Genetics. 2006;173:1813–1816. doi: 10.1534/genetics.106.057927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gresham D, et al. Genome-wide detection of polymorphisms at nucleotide resolution with a single DNA microarray. Science. 2006;311:1932–1936. doi: 10.1126/science.1123726. [DOI] [PubMed] [Google Scholar]
- 16.Toda T, Cameron S, Sass P, Wigler M. SCH9, a gene of Saccharomyces cerevisiae that encodes a protein distinct from, but functionally and structurally related to, cAMP-dependent protein kinase catalytic subunits. Genes Dev. 1988;2:517–527. doi: 10.1101/gad.2.5.517. [DOI] [PubMed] [Google Scholar]
- 17.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 18.Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2003;2:73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- 19.Piper PW. Long-lived yeast as a model for ageing research. Yeast. 2006;23:215–226. doi: 10.1002/yea.1354. [DOI] [PubMed] [Google Scholar]
- 20.Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strauss BS. Cell death and unbalanced growth in Neurospora. J Gen Microbiol. 1958;18:658–669. doi: 10.1099/00221287-18-3-658. [DOI] [PubMed] [Google Scholar]
- 22.Cohen SS, Barner HD. Studies on unbalanced growth in Escherichia coli. Proc Natl Acad Sci USA. 1954;40:885–893. doi: 10.1073/pnas.40.10.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Culbertson MR, Henry SA. Inositol-requiring mutants of Saccharomyces cerevisiae. Genetics. 1975;80:23–40. doi: 10.1093/genetics/80.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henry SA. Death resulting from fatty acid starvation in yeast. J Bacteriol. 1973;116:1293–1303. doi: 10.1128/jb.116.3.1293-1303.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myers CL, et al. Discovery of biological networks from diverse functional genomic data. Genome Biol. 2005;6:R114. doi: 10.1186/gb-2005-6-13-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roosen J, et al. PKA and Sch9 control a molecular switch important for the proper adaptation to nutrient availability. Mol Microbiol. 2005;55:862–880. doi: 10.1111/j.1365-2958.2004.04429.x. [DOI] [PubMed] [Google Scholar]
- 27.Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 28.Zabrocki P, et al. Protein phosphatase 2A on track for nutrient-induced signalling in yeast. Mol Microbiol. 2002;43:835–842. doi: 10.1046/j.1365-2958.2002.02786.x. [DOI] [PubMed] [Google Scholar]
- 29.Kalhor HR, Luk K, Ramos A, Zobel-Thropp P, Clarke S. Protein phosphatase methyltransferase 1 (Ppm1p) is the sole activity responsible for modification of the major forms of protein phosphatase 2A in yeast. Arch Biochem Biophys. 2001;395:239–245. doi: 10.1006/abbi.2001.2558. [DOI] [PubMed] [Google Scholar]
- 30.Wu J, et al. Carboxyl methylation of the phosphoprotein phosphatase 2A catalytic subunit promotes its functional association with regulatory subunits in vivo. EMBO J. 2000;19:5672–5681. doi: 10.1093/emboj/19.21.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan G, Shen X, Jiang Y. Rapamycin activates Tap42-associated phosphatases by abrogating their association with Tor complex 1. EMBO J. 2006;25:3546–3555. doi: 10.1038/sj.emboj.7601239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Como CJ, Arndt KT. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Wang X, Jiang Y. Interaction with Tap42 is required for the essential function of Sit4 and type 2A phosphatases. Mol Biol Cell. 2003;14:4342–4351. doi: 10.1091/mbc.E03-02-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei H, et al. Carboxymethylation of the PP2A catalytic subunit in Saccharomyces cerevisiae is required for efficient interaction with the B-type subunits Cdc55p and Rts1p. J Biol Chem. 2001;276:1570–1577. doi: 10.1074/jbc.M008694200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 36.Dann SG, Selvaraj A, Thomas G. mTOR complex1–S6K1 signaling: at the crossroads of obesity, diabetes and cancer. Trends Mol Med. 2007;13:252–259. doi: 10.1016/j.molmed.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Altomare DA, Testa JR. Perturbations of the AKT signaling pathway in human cancer. Oncogene. 2005;24:7455–7464. doi: 10.1038/sj.onc.1209085. [DOI] [PubMed] [Google Scholar]
- 38.Schacherer J, et al. Genome-wide analysis of nucleotide-level variation in commonly used Saccharomyces cerevisiae strains. PLoS ONE. 2007;2:e322. doi: 10.1371/journal.pone.0000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urban J, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.