Abstract
One-to-one orthologous genes of relatively closely related species are widely assumed to have similar functions and cause similar phenotypes when deleted from the genome. Although this assumption is the foundation of comparative genomics and the basis for the use of model organisms to study human biology and disease, its validity is known only from anecdotes rather than from systematic examination. Comparing documented phenotypes of null mutations in humans and mice, we find that >20% of human essential genes have nonessential mouse orthologs. These changes of gene essentiality appear to be associated with adaptive evolution at the protein-sequence, but not gene-expression, level. Proteins localized to the vacuole, a cellular compartment for waste management, are highly enriched among essentiality-changing genes. It is probable that the evolution of the prolonged life history in humans required enhanced waste management for proper cellular function until the time of reproduction, which rendered these vacuole proteins essential and generated selective pressures for their improvement. If our gene sample represents the entire genome, our results would mean frequent changes of phenotypic effects of one-to-one orthologous genes even between relatively closely related species, a possibility that should be considered in comparative genomic studies and in making cross-species inferences of gene function and phenotypic effect.
Keywords: evolution, mammals, gene essentiality, vacuole
When a species diverges into two separate species, the divergent copies of a single gene in the resulting species are said to be orthologous (1, 2). Although genome-wide patterns of conservation between orthologous genes have been extensively studied at the DNA and protein-sequence levels (3–5) and have started to be investigated at the gene-expression level (6–9), little is known about the evolutionary conservation at the levels of gene function and phenotypic effect on gene deletion. This lack of knowledge is in part due to the widely held presumption that orthologous genes from different species are similar in function and phenotypic effect (2), which probably originated from a few reports that orthologous genes from distantly related species can be swapped without causing apparent phenotypic defects (10–13). Because this presumption is fundamental to comparative genomics (5, 14) and is the basis for using model organisms such as mice to study human biology and disease (15, 16), it deserves a systematic verification.
Two model organisms, the bacterium Escherichia coli (17) and the yeast Saccharomyces cerevisiae (18), have been subject to genome-wide gene-deletion experiments with available information on the fitness of each gene-deletion strain and thus could be compared in terms of the phenotypes of orthologous deletions at the genomic scale. However, these two organisms belong to prokaryotes and eukaryotes, respectively, and are so different even in basic cellular processes that the comparison is neither feasible nor meaningful. We thus choose to compare human (Homo sapiens) and mouse (Mus musculus), which are both placental mammals and have overall similar biology. Our comparison also has practical value because of the common use of mouse as a model organism for studying human biology and disease. In fact, to facilitate the use of mouse models in human biomedical research, the international genetics community recently initiated the Knockout Mouse Project (KOMP) to individually knock out every gene in the mouse genome and acquire phenotypic data (16). Our analysis will be valuable in guiding the proper use of the KOMP data.
In the present study, we focus on one of the most dramatic types of change in a gene's phenotypic effect, namely, a change in gene essentiality. A gene is said to be essential to an organism if the loss of its function renders the fitness of the organism zero; otherwise, the gene is said to be nonessential. We show that >20% of human essential genes have nonessential mouse orthologs and elucidate the mechanisms underlying the changes of gene essentiality in evolution.
Results and Discussion
Many Human Essential Genes Have Nonessential Mouse Orthologs.
From Online Mendelian Inheritance in Man (OMIM) (19), we identified 1,716 human genes with clear gene–disease associations, in which 1,450 genes have unambiguous one-to-one orthologs in the mouse genome (see Methods). This set contains 756 human genes whose mouse orthologs have been experimentally deleted with the resulting phenotypes cataloged in the database of Mouse Genome Informatics (MGI). For the 594 human genes associated with mild diseases, we cannot infer gene essentiality, because mild diseases may be due to mild mutations in essential genes or null mutations in nonessential genes. From the remaining 162 potentially essential genes, we removed 24 immunity-related genes, because the essentiality of their mouse orthologs may not have been adequately assessed in lab. We further removed 18 genes for which there is no evidence that the human disease is due to null mutations. We thus focused on the remaining 120 human genes with clinical features of death before puberty (20) or infertility when null mutations occur, and considered them to be essential in human [supporting information (SI) Dataset S1]. We determined the essentiality of the mouse orthologs of these human genes, based on MGI and relevant literature. Specifically, a mouse gene is considered essential if the knockout mice cannot survive to reproductive age (50 days) or are infertile.
To our surprise, 27 (22.5%) of the 120 mouse orthologs of human essential genes are nonessential (Table 1 and Dataset S1). Furthermore, except for reduced survival or fecundity for Mthfr, Smpd1 Hexb, and Neu1-knockout mice, the other 23 knockout mouse strains (19.2%) are able to breed as successfully as the wild type at least up to the age of 6 months (Table 1). For convenience, we term these 27 human-essential–mouse-nonessential orthologs as HeMn orthologs and the other 93 human-essential–mouse-essential orthologs as HeMe orthologs, where H and M indicate human and mouse, respectively, and the subscripted “e” and “n” indicate essential and nonessential genes, respectively.
Table 1.
One-to-one orthologous genes that are essential in human but nonessential in mouse
| Human gene name | Human disease name | Mouse gene knockout phenotypes |
|---|---|---|
| Arylsulfatase A (ARSA)* | Metachromatic leukodystrophy† | Normal fertility and litter size; impaired balance and spatial learning ability; sulfatide accumulation in the white matter of the brain; reduced myelin sheath thickness in the corpus callosum and optic nerves; a low frequency head tremor develops after 2 years of age. |
| α-Mannosidase, class 2B, member 1 (MAN2B1) | Mannosidosis, α-, types I and II† | Normal development and fertility; no elevated mortality; mild form of human α-mannosidosis |
| Dystrophia myotonica protein kinase (DMPK)* | Myotonic dystrophy-1† | Normal fertility and litter size; abnormal sodium channel gating in cardiac myocytes; cardiac conduction defects; late-onset progressive skeletal myopathy; abnormal muscle intracellular calcium levels |
| Lysosomal acid lipase (LIPA) | Wolman disease; cholesteryl ester storage disease† | Normal development and fertility; accumulation of triglycerides and cholesteryl esters occurs in several organs |
| Axonemal heavy chain dynein type 11 (DNAH11) | Primary ciliary dyskinesia; Kartagener syndrome‡ | Normal fertility; abnormal left–right axis patterning |
| Sialyltransferase 9 (ST3GAL5) | Amish infantile epilepsy syndrome† | Normal viability and fertility; hypoglycemia; increased insulin sensitivity; abnormal lipid level |
| Patched homolog 2 (PTCH2) | Medulloblastoma; basal cell carcinoma† | Normal viability and fertility; normal cell proliferation or differentiation in the cerebellum; abnormal dermal morphology in some males |
| Granulocyte colony-stimulating factor (CSF3R) | Kostmann neutropenia† | Normal development and fertility; reduced numbers of peripheral neutrophils; fewer hematopoietic progenitors in bone marrow; impaired expansion and terminal differentiation of progenitors into granulocytes |
| 5,10-methylenetetrahydrofolate reductase (MTHFR) | Homocystinuria due to MTHFR deficiency† | Reduced survival rate but fertile; delayed development; elevated plasma levels of homocysteine |
| Transforming growth factor-β interacting factor (TGIF1) | Holoprosencephaly-4† | Normal growth, behavior and fertility |
| Aacid phosphatase-2 (ACP2)* | Acid phosphatase deficiency† | Normal development and fertility; skeletal defects in mutants >6 months of age; a small percentage of mutants exhibit tonic–clonic seizures |
| Cathepsin A (CTSA)* | Galactosialidosis† | Normal fertility; death occurs at ≈12 months; aberrant lysosomal storage; enlarged spleen and liver; abormally flat face; reduced body size; generalized edema, ataxia, and tremors |
| N-acetylglucosaminidase (NAGLU)* | Sanfilippo syndrome, type B† | Appear normal, healthy, and fertile up to 6 months of age; survive to 8–12 months; reduced open field activity; massive accumulation of heparan sulfate in kidney and liver; elevated gangliosides in brain; and presence of vacuoles in macrophages, epithelial cells, and neurons. |
| β-Mannosidase (MANBA)* | β-Mannosidosis† | Normal appearance, growth, and fertility to 1 year of age; cytoplasmic vacuolation in central nervous system and visceral organs |
| Ubiquitin–protein ligase e3 component n-recognin 1 (UBR1) | Johanson–Blizzard syndrome† | Normal viability and fertility; 20% lower body weight; reduced muscle and adipose tissue; abnormal metabolism; enhanced nonspatial learning; impaired spatial learning |
| von Willebrand factor-cleaving protease (ADAMTS13) | Atypical hemolytic uremic syndrome; congenital thrombotic thrombocytopenic purpura† | Normal development, viability, and fertility; prolonged vWF-mediated platelet–endothelial interactions |
| Acid sphingomyelinase (SMPD1)* | Niemann–Pick disease, type A and B† | Males could breed until 20 weeks of age and females until 10 weeks of age with normal litter size; life span of 4–8 months; impaired coordination; mild tremor, and ataxia after 8 weeks of age; abnormal lipid homeostasis; decreased body weight |
| β-Glucosidase-1 (GLB1)* | GM1-gangliosidosis; mucopolysaccharidosis IVB† | Normal fertility and litter size; life span of 7–10 months; progressive spastic diplegia; emaciation; accumulation of ganglioside GM1 and asialo GM1 in brain tissue |
| α-1,4-glucosidase (GAA)* | Glycogen storage disease II† | Normal growth and fertility; reduced mobility and strength; impaired coordination, hindlimb paralysis and muscle weakness for the mutants >8 months of age |
| Cytochrome p450, family 7, subfamily b, polypeptide 1 (CYP7B1)* | Giant cell hepatitis, neonatal† | Normal survival, physical appearances, and behaviors; normal bile acid metabolism, plasma cholesterol and triglyceride levels; sterol biosynthetic rates were unaffected in multiple tissues with the exception of the male kidney, which showed an ≈40% decrease |
| Coagulation factor VIII (F8) | Hemophilia A† | Females exhibit normal fertility and pregnancy; males show reduced ability to clot blood; no spontaneous bleeding into joints or soft tissues is observed up to 12 weeks of age |
| Hexosaminidase B (HEXB) | Sandhoff disease† | Normal growth and fertility; mutants exhibit spasticity, muscle weakness, rigidity, tremors, and ataxia beginning ≈4 months of age and resulting in death ≈6 weeks later |
| GM2 activator protein (GM2A)* | GM2-gangliosidosis, AB variant† | Normal growth, survival and fertility; abnormal accumulation of glycolipid and ganglioside in various brain regions with impaired balance, coordination, and learning |
| Very long-chain acyl-CoA dehydrogenase (ACADVL) | Deficiency of Acyl-CoA dehydrogenase, VL† | Normal gross appearance, survival, behavior and fertility; normal body and heart weight at 2 months of age. |
| Alanine:glyoxylate aminotransferase (AGXT) | Hyperoxaluria, primary, type 1† | Normal growth and development; no histological differences between mutants and wild types in multiple tissues; increased oxalate urine levels and higher chance to develop bladder stones for males. |
| Neuraminidase 1 (NEU1)* | Sialidosis, type I and type II† | 27% of the pups in the NMRI background and 10–15% in the C57BL/6 background died suddenly around weaning age; mice that survived past the 21 days were fertile, but stopped producing offspring by the age of 10 weeks; death occurred between the ages of 8 and 12 months. |
| Galactose-1-phosphate uridylyltransferase (GALT) | Galactosemia† | Normal embryonic survival; normal fertility in both sexes; abnormal galactose metabolism, but lack symptoms of acute toxicity. |
**Protein product localized to vacuole.
†Death before puberty.
‡Infertility.
Gene Duplication Is Not the Cause of Gene-Essentiality Changes.
What caused the dramatic change in essentiality between human and mouse in >20% of the examined genes? Previous studies in yeast (21) and nematode (22) suggested that when a gene is deleted, its paralogous gene(s) can often provide functional compensation such that an otherwise essential gene would appear to be nonessential. The HeMn and HeMe orthologs studied here are one-to-one orthologs and hence do not have paralogs that were generated since the human–mouse separation. Nevertheless, it is possible that a paralog that was generated before the human–mouse separation is retained in mouse but lost in human, rendering the effect of functional compensation present in mouse but absent in human. We thus examined whether HeMn-type mouse genes tend to have (i) more paralogs and (ii) closer paralogs in the mouse genome than HeMe-type mouse genes, which could explain why some orthologs of human essential genes are nonessential in mouse. We found that the proportion of mouse genes that have paralogs is not significantly different between the HeMn group (18 of 27 = 66.7%) and the HeMe group (55 of 93 = 59.1%) (P = 0.512, Fisher's exact test). Among the mouse genes that have paralogs, HeMn-type mouse genes do not have significantly more paralogs (average number of paralogs = 4.33) than HeMe-type mouse genes have (average = 3.78) (P = 0.415, Mann–Whitney U test). Moreover, HeMn-type mouse genes are not more similar to their closest paralogs (average protein sequence identity = 56.2%) than HeMe-type mouse genes are to their closest paralogs (average = 58.3%; P = 0.568, U test). Because divergent paralogs are unlikely to compensate one another, we repeated our analysis by considering only paralogs with relatively high protein sequence identities, but our results remain unchanged (Table S1). These observations, consistent with recent reports of a general lack of functional compensation between paralogs in mammals (23, 24), indicate that the dramatic changes of gene essentiality between human and mouse orthologs are not due to differential functional compensation from paralogs. Rather, it is more likely that the evolutionary changes of gene essentiality have resulted from alterations of the genes themselves.
Gene Essentiality Changes Are Associated with Accelerated Protein Sequence Evolution.
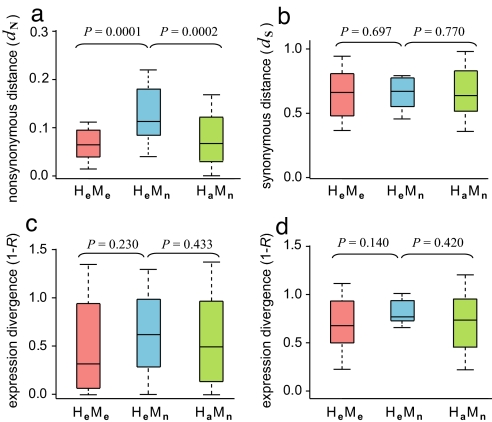
Were changes of gene essentiality more frequently caused by alterations of protein function or gene expression? To address this question, we first estimated the nonsynonymous distance (dN) between each pair of human and mouse orthologs. We found that dN of the HeMn group is significantly greater than that of the HeMe group (P = 5.04 × 10−5, U test) (Fig. 1a), whereas the synonymous distances (dS) are not significantly different between the two groups (P = 0.697, U test) (Fig. 1b). For comparison, let us also define an HaMn group, which includes any human–mouse orthologous pair in which the mouse gene is known to be nonessential. Our HaMn group comprises 864 nonessential non-immune-system mouse genes and their human orthologs. The relatively large dN of the HeMn group compared with that of the HeMe group must be due to accelerated nonsynonymous substitutions caused by (i) weaker purifying selection in the mouse lineage on HeMn genes than on HeMe genes, because mammalian nonessential genes are subject to weaker purifying selection and consequently have higher dN than essential genes (25) and/or (ii) positive selection associated with the change of function and essentiality of HeMn genes. If (i) is the primary reason, the dN of the HeMn group should be lower than that of the HaMn group, because the latter is composed of HnMn and HeMn genes. However, we found that the dN of HeMn genes is significantly greater than that of HaMn genes (P = 1.62 × 10−4, U test) (Fig. 1a), suggesting that (i) cannot be the primary reason of greater dN for HeMn genes than for HeMe genes. Consequently, (ii) must have contributed to a large degree. As expected, there is no significant difference in dS between HeMn and HaMn genes (P = 0.770, U test) (Fig. 1b).
Fig. 1.
The quartile plots of sequence divergence [dN (a) and dS (b)] and expression-profile divergence [ExonArray (c) and GeneAtlas v2 (d)] between human and mouse one-to-one orthologous genes. Values of upper quartile, median, and lower quartile are indicated in each box. The bars indicate semiquartile ranges. H and M indicate human and mouse, respectively, and the subscripts e, n, and a indicate essential, nonessential, and any genes, respectively. The P vales are determined by two-tailed Mann–Whitney U tests.
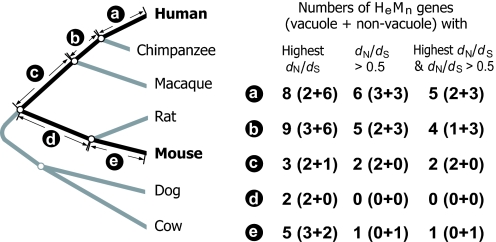
To reconfirm (ii), we used a maximum-likelihood method (26) to estimate branch-specific dN/dS values in a phylogeny of seven placental mammals for each of the 27 HeMn genes (Fig. 2). Besides human and mouse, five additional mammals (chimpanzee, macaque, rat, dog, and cow) were chosen because they can divide the evolutionary path linking human and mouse and because they have publicly available high-quality (i.e., at least 6× coverage) genome sequences so that the orthologous sequences of the HeMn genes can be retrieved. We then compared the dN/dS values for the five branches connecting human and mouse (Fig. 2). An earlier genomic study showed that orthologous genes have, on average, lower dN/dS in rodents than in primates, likely because of a larger population size and consequently increased efficacy of purifying selection in rodents than in primates (27). To make a fair comparison here, we multiplied the estimated dN/dS values for the two rodent branches (d and e in Fig. 2) by 1.23, which is the mean dN/dS value for 5,286 primate genes relative to that for their one-to-one rodent orthologs analyzed in an earlier study (27). Under no positive selection, HeMn genes are expected to exhibit relatively low dN/dS values in branches closer to human and relatively high dN/dS values in branches closer to mouse, along the evolutionary path connecting human and mouse, because essential genes tend to have lower dN/dS than nonessential genes (25). However, we observed the opposite pattern. That is, the fraction of HeMn genes that have their highest dN/dS values in the two branches closest to human (a and b in Fig. 2) is significantly greater than the chance expectation of 2 in 5 (P = 0.014, binomial test). A recent analysis of a high-exchangeability group of amino acid changes suggests that dN/dS > 0.5 likely indicates positive selection (28). Again, we found that the fraction of incidences where dN/dS of an HeMn gene is >0.5 in branch a or b is greater than the chance expectation (P = 0.004, binomial test; Fig. 2). The same is true when both highest dN/dS in a branch and dN/dS > 0.5 are considered (P = 0.015, binomial test; Fig. 2). Taken together, these results suggest that accelerated protein sequence evolution driven by positive selection was associated with changes of gene essentiality in at least an appreciable fraction of HeMn genes and that most HeMn genes had their gene essentiality changed during primate evolution.
Fig. 2.
Variation of branch-specific dN/dS values among the five branches (marked a to e) that connect human and mouse in the mammalian phylogeny. The branch lengths are not drawn to scale. The dN/dS values for branches d and e have been adjusted to correct for the intrinsically low dN/dS in rodents (see Methods).
Gene Expression Evolution Is Not the Cause of Gene Essentiality Changes.
Next, we measured the expression-profile divergence between human and mouse orthologous genes by 1 − R, where R is Pearson's correlation coefficient between their expression levels across homologous tissues of the two species (see Methods). Two independent microarray gene-expression datasets were used. The ExonArray dataset has a higher accuracy in interspecific comparisons (29), whereas the GeneAtlas v2 dataset contains more homologous tissues between the two species (30). Neither dataset shows a significant difference in 1 − R between HeMn genes and HeMe genes (P = 0.230 and 0.140 in Fig. 1 c and d, respectively, U test). Furthermore, 1 − R is not significantly different between HeMn and HaMn genes in these datasets (P = 0.433 and 0.420 in Fig. 1 c and d, respectively, U test). In short, we did not find accelerated gene-expression evolution to be associated with the essentiality changes of HeMn genes. Use of other measures of gene expression divergence gave similar results (Fig. S1).
Gene Essentiality Changes and the Vacuole.
To better understand the biological reasons behind the changes of gene essentiality, we compared the Gene Ontology of the human genes in the HeMn group and the HeMe group using FatiGO (31). There is only one category that is significantly different between the two groups after the control for multiple testing. A much greater fraction of HeMn genes (12 of 27 = 44.4%) than HeMe genes (4 of 93 = 4.3%) have their protein products localized to the vacuole (false discovery rate q = 5.52 × 10−5), a cellular compartment primarily responsible for containing and degrading wastes and toxins. The absence of vacuole proteins in humans tends to cause the accumulation of cellular wastes and toxins that often leads to fatal neurological diseases (Table 1). The mass-corrected basal metabolic rate in human is ≈12% of that in mouse (32), but human reproductive age is ≈150 times that of mouse (Table S2). Consequently, the total amount of waste produced until reproduction for every gram of body mass is ≈18 times higher for human than for mouse. Hence, waste management is much more important in human than in mouse for maintaining proper cellular functions until the time of reproduction. This may have rendered the orthologs of many nonessential mouse vacuole proteins essential in humans. Consistent with this idea, deficiencies of vacuole proteins tend to cause defects at a later life stage in mouse than in human (Table 1). Furthermore, the evolution of the prolonged life history of humans probably generated selective pressures for better vacuole proteins, which may be part of the reason behind the accelerated protein sequence evolution observed in HeMn genes. Comparison of the product of the metabolic rate and the starting reproductive age among primates suggests that the importance of vacuole functions gradually increased in the primate lineage leading to humans, beginning from the common ancestor of all extant primates (Table S2). Consistent with this pattern, HeMn vacuole proteins show accelerated sequence evolution in the three primate branches (a, b, and c) in Fig. 2. However, because of the small sample size, only one comparison yielded statistically significant enrichment in the three branches. That is, for HeMn vacuole proteins, incidences of branch-specific dN/dS > 0.5 occurs more frequently in these three branches than expected by chance (P = 0.028, binomial test).
Approximately 55% of HeMn genes are not vacuole proteins. We confirmed that the results in Fig. 1 remain qualitatively unchanged after the removal of vacuole proteins (Fig. S2). Although the biological reason behind the change of gene essentiality of these nonvacuole proteins is unclear, the association between the essentiality change and accelerated protein-sequence evolution may be similarly caused by an increase in the importance of a particular biological process during human evolution since the human-mouse split, which rendered a nonessential gene essential and at the same time generated selective pressures for the improvement of the gene function. Consistent with this idea, analysis of branch-specific dN/dS indicates that nonvacuole HeMn proteins are significantly more likely to have rapid evolution and highest dN/dS in branch a or b of Fig. 2 than expected by chance (P = 0.002, 0.019, and 0.019, respectively, for the three properties shown in Fig. 2, binomial test).
Final Remarks.
It is possible that the frequency and direction of gene essentiality changes are not the same among evolutionary lineages. For example, the proportion of essential genes in a genome is much greater in mouse than in yeast, which is, in turn, much greater than that in E. coli (24). In the present work, although only HeMn genes are systematically examined, anecdotes of HnMe genes are known. For example, humans with homozygous RECQL null alleles display viable and fertile Bloom's syndrome, whereas targeted deletion of the ortholog in mouse causes embryonic lethality (33). Unfortunately, it is not possible to identify HnMe genes systematically, owing to the difficulty in proving the nonessentiality of human genes. This obstacle notwithstanding, it is almost certain that the prevalence of distinct null phenotypes of human and mouse orthologs is underestimated here. The first reason is that genes with unaltered essentiality could still have altered phenotypic effects. For instance, Adamts2, Acox1, and Fancg are considered essential for human because of the mutant phenotype of premature death (20, 34), but they are essential for mouse because of the knockout phenotype of infertility of adult mice (35–37) (Dataset S1). Second, the phenotypes associated with nonessential genes are probably more labile in evolution than those associated with essential genes, because changes of nonessential genes are expected to be more tolerable than changes of essential genes. Therefore, it is likely that significantly >20% of one-to-one orthologs between human and mouse have different phenotypic effects when deleted. However, we caution that the gene sample analyzed here is relatively small, and thus our results should be reconfirmed when more data become available. In the future, it may also be possible to verify our results by comparing the essentiality of one-to-one orthologous genes from several bacterial species that have been subject to genome-wide gene deletion experiments (38, 17, 39). However, because of high incidences of horizontal gene transfer (40) and nonorthologous gene replacement (41) in prokaryotes, caution should be taken in such comparisons. When studying functional changes in orthologous gene evolution, it is important to distinguish among changes of molecular function, changes of involved biological processes, and changes of physiological importance. By comparing gene essentiality, we are addressing the physiological importance of a gene. A careful examination of Table 1 suggests that the molecular functions and the involved biological processes are likely to be unaltered for the majority of the 27 HeMn genes, whereas their physiological importance has changed dramatically.
Potential implications of our findings are manifold. First, gene annotation based on mutant phenotypes in other species may often be wrong, especially about gene essentiality. Second, comparative and evolutionary analysis depending on the assumption of conservation of gene function or importance between orthologs should be interpreted carefully. Third, alteration of gene essentiality between species could be a cause of the observation that some mutations pathogenic to one species are nevertheless fixed in other species (42, 43). Fourth, it is possible that mouse models of a large number of human diseases will not yield sufficiently accurate information, although they might provide some basic knowledge. The scientific community may need to strategically and systematically consider establishing a primate model organism for studying many human diseases. In this regard, it is particularly important to choose appropriate animal models for the study of human neurological disorders that involve malfunctioning vacuole proteins, because of the opposite essentiality of many vacuole proteins between human and mouse. Finally, the association between changes of gene essentiality and the prolonged life history of humans sheds light on the mechanisms of some human-specific disorders that accompany apparently beneficial human traits.
Although a recent literature survey found otherwise (44), many believe that changes of gene expression are more important than changes of protein function in generating phenotypic differences between species (45, 46). We found that changes of gene essentiality were accompanied by accelerated evolution that was likely driven by positive selection at the protein sequence level but did not find such a signal at the gene-expression level. Although we cannot exclude the possibility that our result regarding expression evolution is caused by the relatively large noise of microarray expression data or the lack of relevant tissues in the datasets analyzed, we can conclude that protein sequence and function changes are important in the change of gene essentiality in evolution. It remains possible, however, that gene expression changes are more important for phenotypic evolution that does not involve a change in gene essentiality.
Methods
Genomic Data and Annotations.
Human genome version NCBI36 and mouse genome version NCBIM36 were used. Annotations of 31,545 human and 28,390 mouse known or predicted genes by Ensembl (release 44) (www.ensembl.org/) were retrieved through BioMart (www.biomart.org/). We considered 14,423 pairs of human–mouse orthologous genes that were annotated as “ortholog_one2one.” This annotation was not based on reciprocal best BLAST hits but was based on phylogenetic analysis (www.ensembl.org/info/about/docs/compara/homology_method.html). The number of synonymous nucleotide substitutions per synonymous site (dS) and the number of nonsynonymous substitutions per nonsynonymous site (dN) between human and mouse orthologs, estimated by the likelihood method, were retrieved from BioMart. The paralog information, including the percentage of sequence identity, was also obtained from BioMart. Because retroduplicates are expected to have unrelated expression patterns from their mother genes and thus are not expected to compensate the loss of the mother genes, we did not consider retroduplicate copies as paralogs of a gene. Retroduplicates were recognized by the absence of introns that are present in their mother genes. Our results remained unchanged when retroduplicates were not excluded.
Human Essential Genes.
There are 1,716 human genes in Ensembl that are associated with heritable human diseases in OMIM (www.ncbi.nlm.nih.gov/omim/). Among them, 1,450 have unambiguous mouse orthologs, and 756 have phenotypic descriptions from gene knockout mice. Following Jimenez-Sanchez and colleagues (20), we categorized essentiality of a gene by the most life-threatening disease that the gene is associated with. Of the 162 human essential genes that have corresponding mouse knockout phenotypes, 24 are immunity related (MP:0005387 in MGI) and were excluded in further analysis, because the sterilized laboratory environment may underestimate the fitness reduction associated with the deletion of immunity genes in mice. To compare with mouse knockout phenotypes, we require that human diseases considered here are due to null (or at least not gain-of-function) mutations. Null mutations are defined as nonsense or frameshift mutations or the absence of gene products in patients as determined by biochemical assays. Eighteen genes were removed because of the lack of evidence for the association between human diseases and null mutations. We manually verified the human and mouse phenotypes for the remaining 120 orthologous genes by reading relevant literature, especially ensuring that the mouse abnormal reproductive system phenotypes annotated in MGI are infertility. The complete list of these 120 genes is provided in Dataset S1.
Mouse Essential and Nonessential Genes.
Mouse phenotypic data were downloaded from MGI version 3.53 (www.informatics.jax.org/). We limited our analysis to null mutants generated by random gene disruption, gene trap mutagenesis, and targeted deletion, together referred to as gene knockout here. Only those genes with one-to-one matches between MGI symbol names and Ensembl gene IDs were kept for subsequent analysis. Genes with phenotypes of embryonic lethality (MP: 0002080), prenatal lethality (MP: 0002081), survival postnatal lethality (MP: 0002082), abnormal reproductive system morphology (MP: 0002160), or abnormal reproductive system physiology (MP: 0001919) were grouped as essential genes. Genes with phenotypes of premature death or induced morbidity (MP: 0002083) were manually inspected and classified according to the literature. If the mutant has a life span of <50 days, the gene is considered to be essential. Genes associated with all other phenotypes (at least MP: 0000001), including the normal phenotype, were grouped as nonessential genes. The dataset included 2,022 essential and 1,655 nonessential mouse genes.
Estimating Branch-Specific dN/dS Values.
Coding sequences (CDS) of HeMn genes from human and mouse and their “one2one” orthologs from chimpanzee (Pan troglodytes), macaque (Macaca mulatta), rat (Rattus norvegicus), cow (Bos taurus), and dog (Canis familiaris) were obtained from BioMart and the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/). If multiple transcripts were annotated for one gene, the longest CDS was chosen. The sequences were aligned by MEGA4 (47) with manual adjustment. Alignment gap sites were subsequently removed. With the known phylogeny of the seven mammals (Fig. 2) (48), the program “codeml” in PAML (49) was used to estimate dN/dS for the HeMn orthologs in each branch of the tree, with the option of “model = 1” chosen in the control file. We then compared the dN/dS values of the five branches (a to e in Fig. 2) that connect human and mouse in the tree. Rodents are known to have intrinsically low dN/dS compared with primates (27). To make a fair comparison of dN/dS among branches, we multiplied the dN/dS estimates for branches d and e by 1.23, which is the mean dN/dS value for 5,286 primate genes relative to that for their one-to-one rodent orthologs (1.28/1.04; see figure S6 of ref. 27).
Microarray Data Analysis.
The GeneAtlas v2 dataset (http://symatlas.gnf.org/) contains the expression data obtained by hybridization of RNAs from 73 human nonpathogenic tissues and 61 mouse tissues onto the Affymetrix microarray chips (human: U133A/GNF1H; mouse: GNF1M) (30). We assigned the probe sets to the human and mouse genes following a previous study (50). The expression level detected by each probe set was obtained as the signal intensity (S) computed from the MAS 5.0 algorithm. The dataset contains 26 common tissues between the two species. They are adipocyte, adrenal gland, amygdala, bone marrow, cerebellum, dorsal root ganglion, heart, hypothalamus, kidney, liver, lung, lymph node, ovary, pancreas, pituitary, placenta, prostate, skeletal muscle, spinal cord, testis, thymus, thyroid, tongue, trachea, trigeminal ganglion, and uterus. Mouse lower spinal cord was used as the homologous tissue of human spinal cord. We measured the expression-profile divergence between a pair of orthologs by 1 − R, where R is Pearson's correlation coefficient between human S and mouse S across the 26 common tissues. We also used another parameter, 1 − ICE, to measure the expression-profile divergence between orthologous genes. ICE (index of coexpression) between two genes is defined as the number of tissues in which both genes are expressed divided by the geometric mean of the number of tissues where each gene is expressed (51). Following convention (30), we used a cutoff of S = 200 to determine whether a gene is expressed in a tissue or not when estimating 1 − ICE.
The ExonArray data were generated by using a microarray platform with more than six million probes targeting all annotated and predicted exons in a genome and were obtained from Xing et al. (29). The data include six common tissues between human and mouse (heart, kidney, liver, muscle, spleen, and testis). We used the expression signals S computed from GeneBASE (http://biogibbs.stanford.edu/∼kkapur/genebase/). The S values were averaged among the three replicated experiments performed for each tissue. Because the quality of the ExonArray data are higher than that of GeneAtlas (29), a cutoff of S = 150 was used to determine whether a gene is expressed in a tissue or not in the estimation of 1-ICE. Under this cutoff, the numbers of genes expressed in a given tissue are similar between the GeneAtlas and ExonArray datasets.
Supplementary Material
Acknowledgments.
We thank Gerardo Jimenez-Sanchez for providing details of the annotation of human genetic diseases. Meg Bakewell, Wendy Grus, David Lipman, Ondrej Podlaha, Wenfeng Qian, and three anonymous referees made valuable comments. This work was supported by a pilot grant from University of Michigan Center for Computational Medicine and Biology and research grants from National Institutes of Health (to J.Z.). B.-Y.L was supported by the University of Michigan Rackham Predoctoral Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800387105/DCSupplemental.
References
- 1.Fitch W. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–106. [PubMed] [Google Scholar]
- 2.Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 3.Li W-H. Molecular Evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 4.Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford Univ Press; 2000. [Google Scholar]
- 5.Koonin E, Galperin M. Sequence—Evolution—Function: Computational Approaches in Comparative Genomics. Boston: Kluwer; 2003. [PubMed] [Google Scholar]
- 6.Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- 7.Jordan IK, Marino-Ramirez L, Koonin EV. Evolutionary significance of gene expression divergence. Gene. 2005;345:119–126. doi: 10.1016/j.gene.2004.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khaitovich P, Enard W, Lachmann M, Paabo S. Evolution of primate gene expression. Nat Rev Genet. 2006;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- 9.Liao BY, Zhang J. Evolutionary conservation of expression profiles between human and mouse orthologous genes. Mol Biol Evol. 2006;23:530–540. doi: 10.1093/molbev/msj054. [DOI] [PubMed] [Google Scholar]
- 10.Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 11.Lutz B, Lu HC, Eichele G, Miller D, Kaufman TC. Rescue of Drosophila labial null mutant by the chicken ortholog Hoxb-1 demonstrates that the function of Hox genes is phylogenetically conserved. Genes Dev. 1996;10:176–184. doi: 10.1101/gad.10.2.176. [DOI] [PubMed] [Google Scholar]
- 12.Acampora D, et al. Murine Otx1 and Drosophila otd genes share conserved genetic functions required in invertebrate and vertebrate brain development. Development. 1998;125:1691–1702. doi: 10.1242/dev.125.9.1691. [DOI] [PubMed] [Google Scholar]
- 13.Nagao T, et al. Developmental rescue of Drosophila cephalic defects by the human Otx genes. Proc Natl Acad Sci USA. 1998;95:3737–3742. doi: 10.1073/pnas.95.7.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mushegian A. Foundations of Comparative Genomics. Burlington, MA: Academic; 2007. [Google Scholar]
- 15.Fox M. The Case for Animal Experimentation: An Evolutionary and Ethical Perspective. Berkeley, CA: Univ of California Press; 1986. [Google Scholar]
- 16.Austin CP, et al. The knockout mouse project. Nat Genet. 2004;36:921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 19.McKusick VA. Mendelian Inheritance in Man. Baltimore: Johns Hopkins Univ Press; 1998. [Google Scholar]
- 20.Jimenez-Sanchez G, Childs B, Valle D. Human disease genes. Nature. 2001;409:853–855. doi: 10.1038/35057050. [DOI] [PubMed] [Google Scholar]
- 21.Gu Z, et al. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- 22.Conant GC, Wagner A. Duplicate genes and robustness to transient gene knock-downs in Caenorhabditis elegans. Proc Biol Sci. 2004;271:89–96. doi: 10.1098/rspb.2003.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang H, Li WH. Gene essentiality, gene duplicability and protein connectivity in human and mouse. Trends Genet. 2007;23:375–378. doi: 10.1016/j.tig.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 24.Liao BY, Zhang J. Mouse duplicate genes are as essential as singletons. Trends Genet. 2007;23:378–381. doi: 10.1016/j.tig.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Liao BY, Scott NM, Zhang J. Impacts of gene essentiality, expression pattern, and gene compactness on the evolutionary rate of mammalian proteins. Mol Biol Evol. 2006;23:2072–2080. doi: 10.1093/molbev/msl076. [DOI] [PubMed] [Google Scholar]
- 26.Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- 27.Gibbs RA, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 28.Tang H, Wu CI. A new method for estimating nonsynonymous substitutions and its applications to detecting positive selection. Mol Biol Evol. 2006;23:372–379. doi: 10.1093/molbev/msj043. [DOI] [PubMed] [Google Scholar]
- 29.Xing Y, Ouyang Z, Kapur K, Scott MP, Wong WH. Assessing the conservation of Mammalian gene expression using high-density exon arrays. Mol Biol Evol. 2007;24:1283–1285. doi: 10.1093/molbev/msm061. [DOI] [PubMed] [Google Scholar]
- 30.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: A web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 32.Tolmasoff JM, Ono T, Cutler RG. Superoxide dismutase: Correlation with life-span and specific metabolic rate in primate species. Proc Natl Acad Sci USA. 1980;77:2777–2781. doi: 10.1073/pnas.77.5.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chester N, Kuo F, Kozak C, O'Hara CD, Leder P. Stage-specific apoptosis, developmental delay, and embryonic lethality in mice homozygous for a targeted disruption in the murine Bloom's syndrome gene. Genes Dev. 1998;12:3382–3393. doi: 10.1101/gad.12.21.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki Y, et al. Peroxisomal acyl CoA oxidase deficiency. J Pediatr. 2002;140:128–130. doi: 10.1067/mpd.2002.120511. [DOI] [PubMed] [Google Scholar]
- 35.Fan CY, et al. Hepatocellular and hepatic peroxisomal alterations in mice with a disrupted peroxisomal fatty acyl-coenzyme A oxidase gene. J Biol Chem. 1996;271:24698–24710. doi: 10.1074/jbc.271.40.24698. [DOI] [PubMed] [Google Scholar]
- 36.Li SW, et al. Transgenic mice with inactive alleles for procollagen N-proteinase (ADAMTS-2) develop fragile skin and male sterility. Biochem J. 2001;355:271–278. doi: 10.1042/0264-6021:3550271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y, et al. Targeted disruption of the murine Fanconi anemia gene, Fancg/Xrcc9. Blood. 2001;98:3435–3440. doi: 10.1182/blood.v98.12.3435. [DOI] [PubMed] [Google Scholar]
- 38.Akerley BJ, et al. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc Natl Acad Sci USA. 2002;99:966–971. doi: 10.1073/pnas.012602299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallagher LA, et al. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc Natl Acad Sci USA. 2007;104:1009–1014. doi: 10.1073/pnas.0606713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 41.Koonin EV, Mushegian AR, Bork P. Non-orthologous gene displacement. Trends Genet. 1996;12:334–336. [PubMed] [Google Scholar]
- 42.Kondrashov AS, Sunyaev S, Kondrashov FA. Dobzhansky–Muller incompatibilities in protein evolution. Proc Natl Acad Sci USA. 2002;99:14878–14883. doi: 10.1073/pnas.232565499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao L, Zhang J. Why are some human disease-associated mutations fixed in mice? Trends Genet. 2003;19:678–681. doi: 10.1016/j.tig.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 44.Hoekstra HE, Coyne JA. The locus of evolution: Evo devo and the genetics of adaptation. Evol Int J Org Evol. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- 45.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 46.Carroll SB. Evolution at two levels: On genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 48.Murphy WJ, Pevzner PA, O'Brien SJ. Mammalian phylogenomics comes of age. Trends Genet. 2004;20:631–639. doi: 10.1016/j.tig.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 49.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 50.Liao BY, Zhang J. Low rates of expression profile divergence in highly expressed genes and tissue-specific genes during mammalian evolution. Mol Biol Evol. 2006;23:1119–1128. doi: 10.1093/molbev/msj119. [DOI] [PubMed] [Google Scholar]
- 51.Lercher MJ, Urrutia AO, Hurst LD. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat Genet. 2002;31:180–183. doi: 10.1038/ng887. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.