Abstract
Microorganisms play a fundamental role in the cycling of nutrients and energy on our planet. A common strategy for many microorganisms mediating biogeochemical cycles in anoxic environments is syntrophy, frequently necessitating close spatial proximity between microbial partners. We are only now beginning to fully appreciate the diversity and pervasiveness of microbial partnerships in nature, the majority of which cannot be replicated in the laboratory. One notable example of such cooperation is the interspecies association between anaerobic methane oxidizing archaea (ANME) and sulfate-reducing bacteria. These consortia are globally distributed in the environment and provide a significant sink for methane by substantially reducing the export of this potent greenhouse gas into the atmosphere. The interdependence of these currently uncultured microbes renders them difficult to study, and our knowledge of their physiological capabilities in nature is limited. Here, we have developed a method to capture select microorganisms directly from the environment, using combined fluorescence in situ hybridization and immunomagnetic cell capture. We used this method to purify syntrophic anaerobic methane oxidizing ANME-2c archaea and physically associated microorganisms directly from deep-sea marine sediment. Metagenomics, PCR, and microscopy of these purified consortia revealed unexpected diversity of associated bacteria, including Betaproteobacteria and a second sulfate-reducing Deltaproteobacterial partner. The detection of nitrogenase genes within the metagenome and subsequent demonstration of 15N2 incorporation in the biomass of these methane-oxidizing consortia suggest a possible role in new nitrogen inputs by these syntrophic assemblages.
Keywords: betaproteobacteria, methanotrophy, nitrogen fixation, syntrophy
Our understanding of the interplay between microorganisms and the environment has advanced tremendously in the past two decades through the application of environmental metagenomics, microanalytical methodologies, novel cultivation methods, and coupling of stable and radiogenic isotopes with molecular analysis of organic biosignatures (i.e., DNA, RNA, protein, and lipids). The strict requirement for pure cultures has been alleviated through the refinement of these techniques, and the field of microbial ecology is now poised to fully illuminate the complex interrelationships within natural communities.
The anaerobic cycling of carbon frequently relies on closely coupled interdependent microbial associations (1). The prevalence and biogeochemical importance of these associations emphasizes the need for effective methodologies that enable the study of novel microbial processes and interspecies relationships in situ. To this end, culture-independent environmental metagenomic studies have expanded our understanding of the phylogenetic and metabolic diversity on Earth (2, 3). An important caveat of these studies is the ability to interpret the metagenomic data in the context of organism identity, which increases in difficulty with increasing community complexity. Direct sequencing of complex soils and sediments, using high-throughput sequencing methods (shotgun cloning or clone-free pyrosequencing), yield important overviews of the metabolic potential of the entire community (4, 5); however, information regarding specific groups of microorganisms and interspecies associations is often lacking. Methods that enable the selective extraction of individual microorganisms from native microbial communities before genome sequencing have shown promise for interpreting genome data in the context of specific microorganisms. Examples of these purification approaches for single cells include microfluidics (6), optical trapping (7), immunomagnetic capture (8), and flow cytometry (9, 10). The ability to identify and characterize the metabolic potential of in situ syntrophic partnerships presents a greater challenge and requires new methodological strategies that are both compatible with metagenomic analysis and can be applied in complex environmental samples, such as anoxic sediments, where these interspecies associations are likely to occur.
In response to this need, we have developed a culture-independent method called “magneto-FISH” that enables the capture and assessment of interspecies associations and corresponding metabolic properties of these organisms directly from complex environments. Briefly, magneto-FISH allows for the targeted magnetic capture of whole microorganisms and cell aggregates, using 16S rRNA oligonucleotide probes to the exclusion of other microorganisms within the environmental sample. In this study, we applied magneto-FISH for targeted metagenomic investigations of in situ syntrophic assemblages, followed by genomics-enabled hypothesis development and subsequent hypothesis testing, using a combination of pyrosequencing and isotope labeling experiments. The magneto-FISH method increases the probability of identifying potentially hidden microbial associations and simplifying environmental genomics by specifically targeting the microorganism(s) of interest.
Here, we investigate a globally significant but poorly understood syntrophic association between as-yet uncultured groups of anaerobic methane-oxidizing archaea (ANME) and sulfate-reducing bacteria; a microbial association responsible for consuming >80% of the methane flux from the ocean (11). We can now begin to address key questions pertaining to the ecology and physiology of these organisms, such as: How versatile is the syntrophic association between specific ANME groups and associated bacteria? What metabolic processes are available to them and how do they specifically acquire essential nutrients in a carbon-dominated environment? Do these properties vary between the different methane-oxidizing ANME ecotypes? Metagenomic studies of ANME archaea have primarily focused on methane and energy metabolism; however, the underlying nutrition and the intricacies of these interspecies associations are still a mystery. Using the phylogenetic selectiveness of the 16S rRNA targeted magneto-FISH assay, we broaden our understanding of the syntrophic lifestyle and specifically nitrogen acquisition of a specific methane-oxidizing ecotype, known as the ANME-2c, and their coassociated bacterial partners recovered from methane-rich deep-sea sediments.
Results and Discussion
Magneto-FISH Technique.
The magneto-FISH method enables the purification of select microorganisms and their physically associated partners directly from environmental samples. Magneto-FISH combines the widely used 16S rRNA-targeted fluorescence in situ hybridization and tyramide signal amplification (CARD-FISH) (12) with the immunomagnetic capture of hybridized cells, using paramagnetic beads coated with an antibody targeting the fluorochrome applied in the CARD-FISH procedure [Fig. 1, supporting information (SI) Fig. S1, and SI Text). Beyond its methodological simplicity, one of the main advantages and important distinctions of magneto-FISH over standard immuno-based capture techniques is a freedom from the requirement of pure cultures and specific antigens synthesized against the target organism. This is especially important for environmental microbiology, where >99% of the known diversity is based on ribosomal sequences (13).
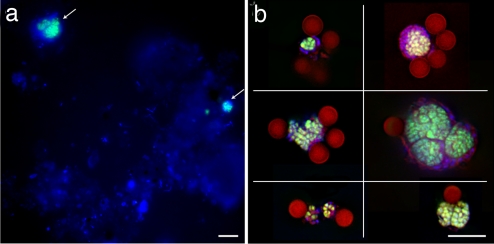
Fig. 1.
CARD-FISH of Eel River Basin sediment with probe ANME-2c_760 (green) and a general DNA stain (DAPI, blue), before (a) and after (b) capture with paramagnetic beads (dark red, collage of six images). (Scale bars, 10 μm.)
Syntrophy and the Anaerobic Oxidation of Methane and Microbial Target Selection for Magneto-FISH.
The history of research on the process of anaerobic oxidation of methane (AOM) has been punctuated with controversy. Once presumed insignificant in nature because of unfavorable thermodynamics and a lack of cultured organisms, we now know that this microbially mediated process is responsible for curbing the majority of methane emissions from the marine environment (reviewed in ref. 11). The unknown microorganisms mediating this process were revealed via a series of molecular and stable isotopic studies over the past decade (ref. 17 and references therein). These uncultured archaeal lineages (known as ANME) are related to methanogens and are believed to catalyze AOM in syntrophic interaction with sulfate-reducing Deltaproteobacteria (14). Substantial diversity exists within the methanotrophic ANME, with three major groups, ANME-1, 2, and 3, currently described (15, 16). In addition, phylogenetically distinct subgroups of the ANME-2 (a, b, and c) appear to exhibit specific habitat preferences and unique spatial configurations with sulfate-reducing bacteria related to Desulfosarcina (DSS) (16, 17), suggesting that the ANME-2 subgroups represent unique ecotypes. Metagenomic sequencing of the microbial community in methane seep sediment has shed light on the physiology and specific mechanism of AOM catalyzed by some of the ANME archaea (18, 19). However, details of the metabolic regulation, environmental adaptation, and intracellular communication of these diverse archaea and their bacterial partners are still largely unexplored. Through the use of magneto-FISH, we extend these earlier investigations to the study of the interspecies consortia as a whole and begin to examine the biochemical underpinnings supporting this complex syntrophic partnership.
We focused our investigation on a specific group of methane oxidizing archaea known as the ANME-2c, using magneto-FISH to selectively capture intact ANME-2c consortia directly from deep-sea methane seep sediments from the Eel River Basin, CA (ERB). The diversity of associated microorganisms and potential metabolism of the ANME-2c associations was subsequently characterized through metagenome sequencing, CARD-FISH, and stable isotope labeling experiments.
Validating the Specificity of the Magneto-FISH Capture: 16S rRNA Analysis and Metagenomics of the Captured ANME-2c Archaea.
The purity of the magneto-FISH captured ANME-2c consortia was evaluated by using both epifluorescence microscopy and DNA extraction, followed by multiple displacement amplification of the genomic DNA and PCR-based screening of ribosomal and metabolic genes (Fig. S2 and ref. 20). Microscopic examination indicated that all captured microorganisms were associated with fluorescently labeled sediment-free ANME-2c aggregates (≈40,000 aggregates captured from 40 μl of sample) (Fig. 1). PCR screening confirmed our CARD-FISH results with ANME-2c phylotypes representing 92% of the total archaeal 16S rRNA sequences recovered from the magneto-FISH captured fraction, compared with 26% in the original sediment (Table 1).
Table 1.
Comparison of 16S rRNA diversity recovered before and after capture of ANME-2c consortia, using Magneto-FISH
| 16S rRNA phylotypes, % |
Closest relative | |
|---|---|---|
| Bulk sediment clone library | Bead capture clone library | |
| Archaea | ||
| 26 | 92 | ANME-2c |
| 6 | 7 | ANME-2a |
| 2 | 1 | Methanococcoides burtonii |
| 19 | 0 | ANME-2b |
| 44 | 0 | Others (ANME-1, MBGD, MBGB) |
| Bacteria | ||
| 25 | 42 | Deltaproteobacteria (Desulfobulbus or Desulfosarcina) |
| 0 | 32 | Betaproteobacteria (Burkholderiaceae) |
| 0 | 9 | Alphaproteobacteria (Sphingomonas) |
| 9 | 2 | Gammaproteobacteria (uncultured) |
| 16 | 7 | Epsilonproteobacteria (uncultured) |
| 0 | 4 | CFB group* (AJ535255, AY768987) |
| 0 | 3 | Planctomyces (AF424488, EF424502) |
| 50 | <1 | Others (eight additional major lineages) |
Archaeal and bacterial 16S rRNA clone libraries were constructed by using Ar20F-958r and Ba27F-1492r, respectively. Total number of clones in archaeal library: 211 (bead capture) and 254 (bulk sediment). Total number of clones in bacterial library: 139 (bead capture) and 165 (bulk sediment).
*CFB group: Cytophaga/Flexibacter/Bacterioides.
A metagenomic screening of the captured ANME-2c assemblage was conducted by using pyrosequencing (454 Life Sciences), yielding 290,073 reads with an length of ≈99 bp each (29 Mb total). Identified protein coding reads were primarily associated with sulfate-reducing Deltaproteobacteria and the ANME-affiliated Methanomicrobiales (see SI Text, Fig. S3, and Table S1). Additionally, this single pyrosequencing run resulted in 24–38% coverage at 1× or greater with verified ANME-2 fosmids (encoding 16S rRNA or methyl coenzyme M reductase, mcrA) previously recovered from the ERB (18) (Fig. S4). As documented in the ANME-1-focused metagenome investigation (18), homologous sequences to all genes in the seven-step methanogenic pathway were recovered (Tables S2 and S3). The dominance of archaeal gene fragments affiliated with the Methanosarcinales in the magneto-FISH captured metagenome, together with the demonstration of ANME-2c enrichment both by PCR and microscopy, demonstrate the specificity and utility of magneto-FISH for targeted metagenomics in complex environmental samples.
Diverse Bacteria Associated with the Methanotrophic Ecotype ANME-2c.
Beyond the capacity to obtain genomic information from a targeted uncultured microorganism (in this case the ANME-2c), a unique feature of the whole-cell magneto-FISH capture is the ability to identify and characterize microorganisms in which the target microorganism directly associate in the environment. Although specific syntrophic associations between the ANME-2 and sulfate-reducing bacteria related to DSS has been well established (14, 16, 17), our findings challenge the specificity of this association by demonstrating tightly coupled physical associations between a single methane-oxidizing ANME subgroup (ANME-2c) and at least three distinct bacterial lineages, including one new sulfate-reducing Deltaproteobacterium and a Betaproteobacterium, not known to be capable of sulfate reduction. This newly discovered bacterial diversity closely associated with the ANME-2c lineage may translate into different ecological strategies or perhaps a broader range of metabolic capabilities expressed by these diverse interspecies associations.
Bacterial 16S rRNA diversity recovered from the magneto-FISH capture was predominantly affiliated with Deltaproteobacteria (55%), including the common syntrophic DSS partner, and an unanticipated second sulfate-reducing bacterial group from the Desulfobulbaceae (Table 1 and Fig. S5). FISH staining of microorganisms in the original seep sediment confirmed the physical association of the ANME-2c ecotype with both DSS and members of the Desulfobulbus, with each individual ANME-2c aggregate associated with either one or the other of these sulfate-reducing groups (Fig. 2a). ANME-2 associations with the Desulfosarcina and Desulfobulbus were observed in approximately equal proportions within the sample. Although representatives of this Desulfobulbus recently have been shown to form associations with another methanotrophic ANME group (ANME-3) (15), relationships between the ANME-2 and Desulfobulbus were not known. These findings illustrate that the diversity of sulfate-reducing bacteria capable of forming partnerships with multiple methanotrophic archaea is greater than previously recognized. Also, and perhaps more indicative of the versatility of the AOM syntrophy, is the observed variety in bacterial partnerships that can be supported by a single ANME ecotype (ANME-2c) within a localized environment.
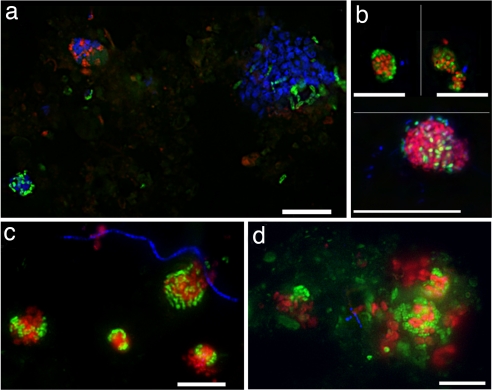
Fig. 2.
Fluorescence in situ hybridization (CARD-FISH) of Eel River Basin sediment. (a and b) Shown are oligonucleotide probes targeting archaeal ANME-2c (ANME2c_760, blue), Desulfosarcina (DSS_225, green), and Desulfobulbaceae (DBB_660, red) (a) and ANME-2 archaea (Eel_932, red), Desulfobacteriaceae (DSS_658, green), and Betaproteobacteria (Bet_42a, blue) (b Lower). Collage of two pictures. (b Upper) ANME-2 archaea (Eel_932, red) and Betaproteobacteria (Bet_42a, green) and a general DNA stain (DAPI, blue). (c and d) ANME-2 (Eel_932, red), Desulfobacteriaceae (DSS_658, green), and Alphaproteobacteria (Alpha_986, blue). (Scale bars, 10 μm.)
The diversity in physically associated bacteria with the ANME-2c ecotype, documented by both FISH and 16S rRNA gene analysis, leads to fundamental questions regarding the physiology and metabolism of the two distinct groups of Deltaproteobacteria and of the newly described Betaproteobacteria. Neither of the sulfate-reducing syntrophic lineages have been cultured; however, there are characteristic distinctions in the carbon mineralization pathway for the Desulfobacteraceae and Desulfobulbaceae. Species belonging to Desulfosarcina/Desulfococcus (Desulfobacteraceae) are capable of complete oxidation using the carbon monoxide dehydrogenase pathway (CODH), whereas members of the Desulfobulbaceae are typically incapable of complete conversion of carbon to CO2 (21). These metabolic differences could ultimately alter the terminal products and energy gained by the two distinct syntrophic assemblages assuming similar electron transfer equivalents for both associations. The implications of these differences may impact the growth efficiency and the stoichiometry of end products generated per mole of methane by the Desulfosarcina/ANME-2c and Desulfobulbus/ANME-2c consortia.
Predictions regarding common substrates shared between the two sulfate-reducing genera that may serve as the intermediate in AOM are less obvious. A broad comparison of substrate profiles between cultured members of the Desulfobacteraceae and Desulfobulbaceae show that the metabolism of ethanol, propionate, lactate, and hydrogen are common for both families (21). Hydrogen has been frequently discussed as the possible intermediate in the AOM syntrophy (22); however, experiments amended with H2 show little to no effect on AOM (23, 24). It is possible that the electron transfer agent is distinct from substrates commonly tested with lab cultures. For example, recent evidence from incubation experiments invokes the transfer of methyl sulfides between methanotrophic archaea and sulfate-reducing bacteria (24). In Moran et al. (24), representatives of Desulfobulbus were one of the sulfate-reducing bacterial groups detected in enrichments from methane seep sediment supplemented with methanethiol. Other possibilities, such as electron shuttles (e.g., anthraquinone-2,6-disulphonate and phenazines), have been explored but not thoroughly tested (25).
Results from the magneto-FISH capture suggest the bacterial diversity involved in the ANME-2c association may extend beyond sulfate-reducing Deltaproteobacteria (Table 1 and Fig. S5). In particular, Betaproteobacteria, most similar to members of the Burkholderiaceae, and Alphaproteobacteria, related to Sphingomonas, comprised a significant fraction of the bacterial sequences within both the PCR-based 16S rRNA survey (32% and 9% total, respectively; Table 1) and protein coding bacterial pyrosequencing reads (668 and 1,044 reads of 20,083; Figs. S3 and S6). Other phylotypes comprising 2–7% of the recovered bacterial sequences within the ANME-2c library were related to organisms common within the methane seep environment, including Gamma and Epsilonproteobacteria, Planctomycetes, and the CFB group (Table 1 and Fig. S5). The association of the Alpha and Betaproteobacteria with the ANME-2c ecotype was confirmed with FISH. Triple CARD-FISH, using either an Alphaproteobacteria- or Betaproteobacteria-specific probe, a probe targeting the Desulfobacteriaceae combined with probes targeting both the ANME-2 or the specific subclade, ANME-2c, confirmed the presence of Alphaproteobacterial and Betaproteobacterial lineages in association with ANME-2 consortia (Fig. 2). In some instances (<1% total cell aggregates), Betaproteobacteria were observed as the dominant or sole bacterial partner associated with the ANME-2 (Fig. 2b), whereas in other cases, a few cells targeted with the Alphaproteobacterial or Betaproteobacterial probes were observed to be loosely associated on the periphery of the ANME-2c/DSS aggregates (Fig. 2 c and d).
Betaproteobacteria are common in freshwater and soil biomes but not frequently described as being from the marine environment. 16S rRNA gene surveys from hydrate-bearing deep subsurface sediments and methane seeps have reported Burkholderiaceae-related phylotypes (26), suggesting a potential niche for these organisms within methane-rich marine environments; however, their role in this ecosystem remains undefined. There has been some speculation that Betaproteobacteria may participate in methanotrophy (27); however, we are unaware of another study describing a close physical association between these organisms and ANME archaea. The involvement of Betaproteobacteria within the Chlorochromatium symbiosis, and other documented couplings between Betaproteobacteria and bacterial methanotrophs (28) suggest some conferred capacity for interspecies associations by this proteobacterial subdivision. The associations visually documented between Betaproteobacteria and the ANME-2c within the ERB methane seeps were rare compared with associations with Deltaproteobacteria, which may explain their lack of detection in previous studies. Independent of their abundance, the documentation of this association serves to illustrate the diversity of interspecies interactions maintained by a single lineage of methanotrophic archaea.
Preliminary data from the metagenome hints at a potentially broader range of metabolic strategies conferred by these organisms. Within the ANME-2c metagenome, ≈3% of the bacteria-associated protein coding reads were affiliated with betaproteobacteria (Fig. S3), with up to 28% orthologous reads identified in comparisons against completed genomes from the Burkholderiaceae (Table S1). An initial analysis of this data suggests that denitrification may be one possible metabolic strategy conferred by the Betaproteobacteria, a hypothesis based on the identification of gene fragments related to membrane bound dissimilatory nitrate reductase (narG and narH) and nitric oxide reductase (norB) (Table S4). Additional sequences related to nitrite reductase (nirB) and nitrous oxide reductase (nosZ) from other bacterial groups were also recovered. In light of the recent discovery of methane oxidation coupled to nitrate reduction by archaeal-bacterial consortia in freshwater systems (29), this observation may suggest a possible role for denitrification in the methane seep environment. The variable concentrations of NO3− measured within the seep ecosystem, such as sites affiliated with filamentous sulfide-oxidizing bacteria (30), illustrate the tremendous geochemical heterogeneity occurring within this environment, which may contribute to the diversity in interspecies associations observed. Although the direct assignment of genes to Betaproteobacteria and determination of denitrification activity in situ requires confirmation, the observations developed through this targeted genomic overview of the syntrophic association aids in the identification and framing of new research directions.
Testing Metagenomic Predictions of Nitrogen Fixation with Stable Isotope Labeling Experiments.
The links between the carbon, nitrogen, and sulfur cycle within methane-impacted deep-sea environments are currently not well defined. In particular, a comprehensive nitrogen budget for these environments is needed. Nitrogen fixation has been postulated to play a role in the input of new nitrogen to these communities; however, diazotrophic microorganisms within the methane seep environment have not been identified. Prior investigations have suggested that many seep communities may be nitrogen limited, based on δ15N depleted total organic nitrogen values and increased CH4 oxidation resulting in skewed Redfield ratios for pore water dissolved inorganic carbon and nitrogen (30, 31). Perhaps exacerbating the problem, microbial biomass sustained within the methane seep can be an order of magnitude greater than surrounding deep-sea sediments (17), resulting in increased nitrogen demand. Concentrations of pore water ammonium within the ERB methane seeps vary up to 10-fold, with an average value of ≈40 μM (S. Joye and M. Boles, personnel communication). Nitrate concentrations within distinct methane seep habitats are also likely dynamic, in large part driven by nitrate-respiring Beggiatoa (30), creating additional spatial and temporal variation in nitrogen availability.
The metagenomic data raises various hypotheses about the involvement of the ANME-2c assemblage in new nitrogen acquisition within this highly productive deep-sea habitat. The recovery of nitrogenase gene fragments and other genes presumably involved in nitrogen fixation, related to both Deltaproteobacteria and Methanosarcina in our metagenome data suggests a potential role for the ANME-2c consortia in new nitrogen production within methane seeps (Table S5). The phylogeny and diversity of these putative diazotrophs was further defined with PCR targeted assays for the nifH subunit from both the magneto-FISH captured consortia and from bulk DNA extracts from methane seep sediment. Diverse nitrogenase genes were recovered from both the captured ANME-2c DNA and environmental DNA, primarily associated the nifH Group III (32), which includes diazotrophic sulfate-reducing bacteria and methanogens (Fig. 3). The detection of nifH from other methane seep areas in the ERB suggests that the genetic potential for nitrogen fixation may be common in this environment. Further indication that members of the methane-oxidizing consortia may be directly participating in nitrogen fixation is supported by a recent unpublished GenBank submission of a near complete nitrogenase operon putatively affiliated with methanotrophic archaea from methane seeps off of Japan (GenBank accession no. AB362194). This sequence shows a high degree of relatedness to identified nitrogenase gene fragments recovered in our ANME-2c metagenome library.
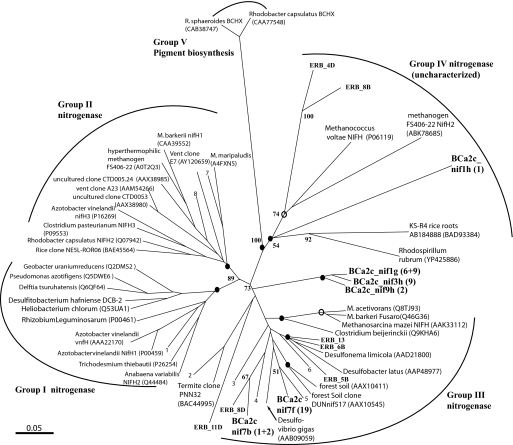
Fig. 3.
Unrooted neighbor-joining (NJ) tree for translated nifH sequences. The topology of the NJ tree is consistent with phylogeny produced by using Bayesian analysis (MrBayes), with the exception of Eel River Basin clone ERB_11D, placed as a well supported deep lineage in nifH cluster II. Bootstrap support values were obtained from neighbor joining (1,000 resamplings). Solid circles represent nodes with posterior probability support (>70%) determined by Bayesian analysis (MrBayes running mixed model with 1 × 106 resamplings). Posterior probabilities between 50% and 70% are indicated by open circles. Taxa listed as numbers in the tree are as follows: 1, Uncultured CB895H9 (AAO67617); 2, Termite clone CfN-RT12 (BAC21035); 3, Rice Cluster I methanogen (CAJ36850); 4, delta-proteobacteria MLMS1 (ZP 01287662); 5, uncultured Diazotroph (AAM14782); 6, Syntrophobacter fumaroxidans MPOB (YP845148); 7, Methanobacterium thermoautotrophicus (AAP48984); and 8, Methanobacterium thermolithotrophicus (CAA32055). (Scale bar, 0.05 changes per amino acid position.)
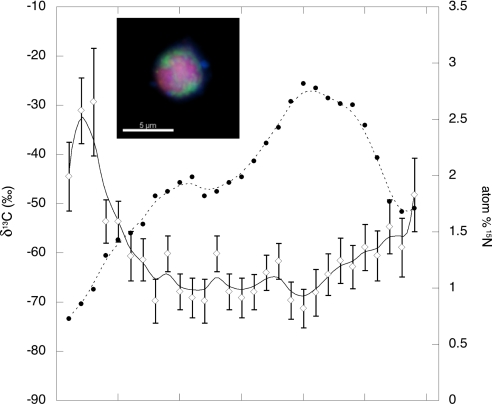
The potential for diazotrophy by the methane-oxidizing consortia is intriguing considering the predicted low free energy yield gained through the syntrophic sulfate-methane redox couple (23, 33). Nitrogen fixation is one of the most energetically expensive anabolic processes, requiring a significant cellular investment in both ATP and reducing equivalents (34). Preliminary 15N2 incubation experiments designed to test for nitrogen fixation by the consortia within the ERB methane seep sediments were conducted by using a combination of FISH and secondary ion mass spectrometry (FISH-SIMS) (SI Text and ref. 24). Incubations of methane seep sediments were over pressured with a headspace of 5% 15N2 (99.8%) and a balance of methane (2 atm) and incubated in the dark at 8°C. Samples used for FISH-SIMS analysis were collected after a 6-month incubation period, enabling sufficient time to detect the 15N label in the biomass of the slow growing ANME aggregates [≈1–2 doublings (25, 35)] (V.J.O. and C. House, unpublished data). SIMS analysis of individual archaeal-bacterial aggregates with light δ13C values (below −50‰), suggestive of methanotrophic growth, revealed 2–3% enrichment in 15N of total cellular nitrogen. The greatest enrichment in 15N appeared to be associated with methanotrophic archaea, interpreted from the SIMS generated depth profile recording both natural abundance δ13C and fractional abundance 15N (atom %) through a layered archaeal-bacterial aggregate (Fig. 4). Fig. 4 illustrates that the greatest 15N incorporation (2.8%) corresponds with the light δ13C core of methane-oxidizing archaea, with values down to −71.2‰. For comparison, another aggregation of cells with more 13C-enriched biomass (−36‰) contained comparably less 15N (0.6 atom %). This data supports the genomic prediction of N2 fixation potential, demonstrating diazotrophy is possible within deep-sea methane seeps and suggesting that methanotrophic consortia are linked to this process. The significance of N2 fixation to the nutrition of methanotrophic consortia in situ remains to be determined. Although the nitrogen budget within the methane seep ecosystem is not well understood, it is reasonable to assume that the inherent heterogeneity likely results in spatial and temporal variability of nitrogen supply within these dynamic, high productivity habitats.
Fig. 4.
FISH-SIMS acquired profile of carbon-13 and nitrogen-15 within a layered archaeal-bacterial cell aggregate (Inset) recovered from a sediment incubation amended with a headspace containing 15N-dinitrogen and methane. Epifluorescence image of aggregate is shown in Inset, where archaea are depicted in red and bacteria in green. Both natural abundance δ13C (‰) of the archaeal/bacterial aggregate (open diamonds) and fractional abundance (%) 15N (solid circles), representing the amount of 15N label incorporated into aggregate biomass, are depicted. For δ13C values, analytical precision (1σ) is shown. The x axis represents increasing time of Cs+ ion beam exposure during the analysis from left to right (arbitrary units), where aggregate cell material is progressively sputtered away over the course of the analysis. This results in a cross-section profile of 13C and 15N, where values for the outer cell layers (bacteria) are shown on the left and right most portion of the graph, transitioning through the methane-oxidizing archaeal core in the center.
Conclusion
Magneto-FISH allows for the recovery of low diversity metagenomic sequences that can be attributed directly to the target microorganism and their physically associated partners within complex environments. This new methodological strategy provides a valuable roadmap for generating testable hypotheses regarding how specific microorganisms and syntrophic consortia operate in nature. The application of magneto-FISH to uncultured methane-oxidizing consortia from deep-sea methane seeps demonstrated a greater diversity among the associated members than previously recognized. Whether these different microbes are integral to the syntrophy or merely physically associated but not actively involved warrants further research. The documentation of these diverse associations and the potential implications for versatile metabolic interactions by a single ANME subgroup or ecotype is significant both from the context of the ecology and evolution of this putatively ancient form of metabolic cooperation. The potential for metabolic versatility, a trait common to cultured methanogenic relatives of the ANME-2 (36), combined with the ability to form diverse syntrophic associations may be the secret to the successful distribution of this biogeochemically significant group of microorganisms. Further studies building on the observations, such as N2 fixation, and hypotheses relating to other metabolic strategies, such as denitrification, possibly conferred by these organisms will be essential for assessing the broader ecological role of the methanotrophic ANME consortia. This ultimately may aid in understanding the complex biological controls of methane flux by these cooperative microbial partnerships in the deep-sea.
Materials and Methods
Magneto-FISH: Fluorescence in Situ Hybridization for Subsequent Paramagnetic Bead Capture (37).
To separate paraformaldehyde fixed microorganisms from sediment particles, two 1-ml subsamples were each mixed with 35 ml of 1× PBS (see SI Text) containing 0.01 M sodium pyrophosphate and sonicated on ice with a sonicating wand (Branson sonifier 150) for two 10-s pulses at 8 W. The slurry was centrifuged at 1,000 × g for 5 min at 4°C, resuspended in 2 ml of equal parts 1× PBS:ethanol, spun again at 500 × g, resuspended in 2 ml of absolute ethanol and stored at −20°C. For permeabilization of the cells and removal of external minerals associated with the cell aggregates, a 40-μl subsample of sonicated sediment was added to 100 ml of 1× TE [10 mM Tris·HCl and 1 mM EDTA (pH 9.0)] and treated in a histological microwave oven (Microwave Research and Applications) at 65°C for 2 min. This treatment did not appear to disrupt or lyse the fixed cell aggregates. Subsequently, the slurry was placed on ice and centrifuged for 5 min at 1,000 × g at 4°C. To inactivate endogenous peroxidases, the pellet was resuspended in 100 ml of 1× PBS containing 0.1% H2O2, vortexed, kept at room temperature (RT) for 1 min, and centrifuged at 1,000 × g for 4 min. The pellet was resuspended in 2 ml of hybridization buffer containing 60% formamide (vol/vol) and 0.5 ng·μl−1 of horseradish peroxidase labeled oligonucleotide probe targeting ANME-2c (ANME_2c_760) (17). This mix was then added to a 2-ml reaction tube, placed into a 100-ml water bath and heated at 46°C for 30 min at a power output of 50% (microwave oven). Unbound probe was removed by mixing the hybridized sediment with 100 ml of 1× PBS and incubating at RT for 10 min, followed by centrifugation of sample at 1,000 × g for 5 min at 4°C. As a negative control, sediment was hybridized with an HRP-labeled nonsense-probe, NON338 (38), which did not produce a CARD-FISH signal in our samples.
Catalyzed Reporter Deposition.
The sediment pellet was resuspended in 5 ml of amplification buffer containing 0.0015% H2O2 and 0.5 μg of each fluorescein- and biotin-labeled tyramide (both custom labeled) according to ref. 39 and incubated at 37°C for 15 min. The biotin labeled tyramide served to enhance the signal intensity, because we observed a brighter and pH-independent CARD-FISH signal in the presence of biotin-tyramide (37). The hybridized sediment was mixed with 100 ml of 1× PBS, centrifuged at 1,000 × g for 5 min, resuspended in 100 ml of 1× PBS containing 0.5% BSA (BSA) and microwaved at 40°C for 20 min to remove the unbound tyramide. After the final wash, the sample was collected by centrifugation at 1,000 × g for 5 min, and resuspended in 2 ml of 1× PBS containing 0.01 M sodium pyrophosphate and sonicated on ice for 5 s to dislodge cells and cell aggregates from sediment particles.
Cell Capture with Paramagnetic Beads.
The CARD-FISH stained sediment sample was split into two 1-ml portions, 200 μl of BSA, and 100 μl of monoclonal mouse anti-fluorescein-antibody (Molecular Probes)-labeled pan-mouse paramagnetic beads (5 μm diameter) (Dynal) were added to each portion, and both were incubated at RT for 1 h on a rotator. To obtain a pure fraction of hybridized target cells free from sediment particles and unlabeled cells, an apparatus consisting of two separation funnels was used (Fig. S1). Briefly, the lower funnel (A) was filled with PBS buffer and the sediment slurry after incubation with the paramagnetic beads was gently pipetted into funnel A. A neodymium ring magnet placed around the neck of funnel A was used to concentrate the paramagnetic beads and captured target cells, and the nonmagnetic residue was separated by gravity. Gentle washing of the magneto-FISH captured cells was accomplished by using a second reservoir filled with PBS (funnel B) seated on top of funnel A. To recover the paramagnetic beads and captured cells, the magnet was removed, the funnel was tilted, and the captured cells were resuspended in a small volume of 1× TE (0.5 ml) and drained through the top opening of funnel A. A second wash step with 0.5 ml of 1× TE was used to recover remaining cells in funnel A and pooled with the first. A detailed description of this procedure is provided in SI Text.
Supplementary Material
Acknowledgments.
We thank C. House for assistance with the ion microprobe measurements; D. Newman, J. Grotzinger, M. Joye, C. Gammon, D. Fike, I. Head, and two anonymous reviewers for invaluable comments that improved this manuscript; and S. Johnson, F. Rohwer, R. Edwards, B. Orcutt, and the 2006 science party of cruise AT 15–11 and pilots of the D.S.R.V. Alvin for their assistance with various aspects of this work. This work was supported by National Science Foundation Award Grant MCB-0348492, the Gordon and Betty Moore Foundation, a Davidow grant to Caltech's GPS Division (to V.J.O.); National Institutes of Health Grant P50 HG004071 (to C.T.B.), in part by a Caltech GPS Texaco fellowship (to A.P.), and a National Science Foundation graduate fellowship (to A.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [accession nos. EU622281–EU622312 (16S rRNA) and EU647340–EU647354 (nifH)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0711303105/DCSupplemental.
References
- 1.Schink B. Synergistic interactions in the microbial world. Antonie van Leeuwenhoek. 2002;81:257–261. doi: 10.1023/a:1020579004534. [DOI] [PubMed] [Google Scholar]
- 2.Tyson GW, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 3.DeLong E, Karl D. Genomic perspectives in microbial oceanography. Nature. 2005;437:336–342. doi: 10.1038/nature04157. [DOI] [PubMed] [Google Scholar]
- 4.Edwards RA, et al. Using pyrosequencing to shed light on deep mine microbial ecology under extreme hydrogeologic conditions. BMC Genomics. 2006;7:57. doi: 10.1186/1471-2164-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 6.Marcy Y, et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci USA. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashkin A. Optical trapping and manipulation of neutral particles using lasers. Proc Natl Acad Sci USA. 1997;94:4853–4860. doi: 10.1073/pnas.94.10.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeung YA, Wittrup KD. Quantitative screening of yeast surface-displayed polypeptide libraries by magnetic bead capture. Biotechnol Prog. 2002;18:212–220. doi: 10.1021/bp010186l. [DOI] [PubMed] [Google Scholar]
- 9.Kalyuzhnaya MG, et al. Fluorescence in situ hybridization-flow cytometry-cell sorting based method for separation and enrichment of type I and type II methanotroph populations. Appl Environ Microbiol. 2006;72:4293–4301. doi: 10.1128/AEM.00161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sekar R, Fuchs BM, Amann RI, Pernthaler J. Flow sorting of marine bacterioplankton after fluorescence in situ hybridization. Appl Environ Microbiol. 2004;70:6210–6219. doi: 10.1128/AEM.70.10.6210-6219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reeburgh WS. Oceanic methane biogeochemistry. Chem Rev. 2007;107:486–513. doi: 10.1021/cr050362v. [DOI] [PubMed] [Google Scholar]
- 12.Pernthaler A, Pernthaler J, Amann R. Fluorescence in situ hybridization and catalyzed reporter deposition (CARD) for the identification of marine bacteria. Appl Environ Microbiol. 2002;68:3094–3101. doi: 10.1128/AEM.68.6.3094-3101.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boetius A, et al. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature. 2000;407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- 15.Niemann H, et al. Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature. 2006;443:854–858. doi: 10.1038/nature05227. [DOI] [PubMed] [Google Scholar]
- 16.Orphan VJ, et al. Comparative analysis of methane-oxidizing archaea and sulfate- reducing bacteria in anoxic marine sediments. Appl Environ Microbiol. 2001;67:1922–1934. doi: 10.1128/AEM.67.4.1922-1934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knittel K, et al. Diversity and distribution of methanotrophic Archaea at cold seeps. Appl Environ Microbiol. 2005;71:467–479. doi: 10.1128/AEM.71.1.467-479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hallam SJ, et al. Reverse methanogenesis: Testing the hypothesis with environmental genomics. Science. 2004;305:2004. doi: 10.1126/science.1100025. [DOI] [PubMed] [Google Scholar]
- 19.Meyerdierks A, et al. Insights into the genomes of archaea mediating the anaerobic oxidation of methane. Environ Microbiol. 2005;7:1937–1951. doi: 10.1111/j.1462-2920.2005.00844.x. [DOI] [PubMed] [Google Scholar]
- 20.Binga EK, Lasken RS, Neufield JD. Something from (almost) nothing: The impact of multiple displacement amplification on microbial ecology. ISME J. 2008:1–9. doi: 10.1038/ismej.2008.10. [DOI] [PubMed] [Google Scholar]
- 21.Rabus R, Hansen T, Widdel F. Dissimilatory sulfate- and sulfur-reducing prokaryotes The Prokaryotes. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. Vol 2. New York: Springer; 2006. pp. 659–768. [Google Scholar]
- 22.Hoehler TM, Alperin MJ, Albert DB, Martens CS. Field and laboratory studies of methane oxidation in an anoxic marine sediment: Evidence for a methanogenic-sulfate reducer consortium. Global Biogeochem Cycles. 1994;8:451–463. [Google Scholar]
- 23.Nauhaus K, Boetius A, Krüger M, Widdel F. In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate area. Environ Microbiol. 2002;4:296–305. doi: 10.1046/j.1462-2920.2002.00299.x. [DOI] [PubMed] [Google Scholar]
- 24.Moran JJ, et al. Methyl sulfides as intermediates in the anaerobic oxidation of methane. Environ Microbiol. 2008;10:162–173. doi: 10.1111/j.1462-2920.2007.01441.x. [DOI] [PubMed] [Google Scholar]
- 25.Nauhaus K, Treude T, Boetius A, Krüger M. Environmental regulation of the anaerobic oxidation of methane: A comparison of ANME-I and ANME-II communities. Environ Microbiol. 2005;7:98–106. doi: 10.1111/j.1462-2920.2004.00669.x. [DOI] [PubMed] [Google Scholar]
- 26.Lanoil BD, et al. Bacteria and Archaea physically associated with Gulf of Mexico gas hydrates. Appl Environ Microbiol. 2001;67:5143–5153. doi: 10.1128/AEM.67.11.5143-5153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchesi JR, et al. Methanogen and bacterial diversity and distribution in deep gas hydrate sediments from the Cascadian Margin as revealed by 16S rRNA molecular analysis. FEMS Microbial Ecol. 2001;34:221–228. doi: 10.1111/j.1574-6941.2001.tb00773.x. [DOI] [PubMed] [Google Scholar]
- 28.Bothe H, et al. Heterotrophic bacteria growing in association with Methylococcus capsulatus (Bath) in a single cell protein production process. Appl Microbiol Biotechnol. 2002;59:33–39. doi: 10.1007/s00253-002-0964-1. [DOI] [PubMed] [Google Scholar]
- 29.Raghoebarsing AA, et al. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature. 2006;440:918–921. doi: 10.1038/nature04617. [DOI] [PubMed] [Google Scholar]
- 30.Joye SB, et al. The anaerobic oxidation of methane and sulfate reduction in sediments from Gulf of Mexico cold seeps. Chem Geol. 2004;205:219–238. [Google Scholar]
- 31.Valentine DL, et al. Biogeochemical investigations of marine methane seeps, Hydrate Ridge, Oregon. J Geophys Res. 2005;110:G02005. [Google Scholar]
- 32.Raymond J, Siefert JL, Staples CR, Blankenship RE. The natural history of nitrogen fixation. Mol Biol Evol. 2004;3:541–554. doi: 10.1093/molbev/msh047. [DOI] [PubMed] [Google Scholar]
- 33.Valentine DL. Biogeochemistry and microbial ecology of methane oxidation in anoxic environments: A review. Antonie van Leeuwenhoek. 2002;81:271–282. doi: 10.1023/a:1020587206351. [DOI] [PubMed] [Google Scholar]
- 34.Howard JB, Rees DC. How many metals does it take to fix N2? A mechanistic overview of biological nitrogen fixation. Proc Natl Acad Sci USA. 2006;103:17088–17093. doi: 10.1073/pnas.0603978103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Girguis PR, Cozen AE, DeLong E. Growth and population dynamics of anaerobic methane-oxidizing archaea and sulfate-reducing bacteria in a continuous-flow bioreactor. Appl Environ Microbiol. 2005;71:3725–3733. doi: 10.1128/AEM.71.7.3725-3733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galagan JE, et al. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 2002;12:532–542. doi: 10.1101/gr.223902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pernthaler A, Orphan VJ. 11/746,374. US Patent. 2007
- 38.Manz W, et al. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: Problems and solutions. System Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 39.Pernthaler A, Pernthaler J, Amann R. In: Molecular Microbial Ecology Manual. Kowalchuk G, de Bruijn FJ, Head IM, Akkermans ADL, van Elsas JD, editors. The Netherlands: Kluwer Academic, Dordrecht; 2004. pp. 711–726. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.