Abstract
Jasmonate (JA) is a lipid-derived hormone that regulates diverse aspects of plant immunity and development. An amino acid-conjugated form of JA, jasmonoyl–isoleucine (JA–Ile), stimulates binding of the F-box protein coronatine-insensitive 1 (COI1) to, and subsequent ubiquitin-dependent degradation of, jasmonate ZIM domain (JAZ) proteins that repress transcription of JA-responsive genes. The virulence factor coronatine (COR), which is produced by plant pathogenic strains of Pseudomonas syringae, suppresses host defense responses by activating JA signaling in a COI1-dependent manner. Although previous data indicate that COR acts as a molecular mimic of JA–Ile, the mechanism by which JA–Ile and COR are perceived by plant cells remains unknown. Here, we show that interaction of tomato COI1 with divergent members of the JAZ family is highly specific for JA–Ile and structurally related JA conjugates and that COR is ≈1,000-fold more active than JA–Ile in promoting this interaction in vitro. JA–Ile competes for binding of COR to COI1–JAZ complexes, demonstrating that COR and JA–Ile are recognized by the same receptor. Binding of COR to the COI1–JAZ complex requires COI1 and is severely impaired by a point mutation in the putative ligand-binding pocket of COI1. Finally, we show that the C-terminal region of JAZ3 containing the highly conserved Jas motif is necessary and sufficient for hormone-induced COI1–JAZ interaction. These findings demonstrate that COI1 is a critical component of the JA receptor and that COR exerts its virulence effects by functioning as a potent agonist of this receptor system.
Keywords: plant immunity, ubiquitin, JAZ protein, plant defense, jasmonic acid
Jasmonate (JA) and its bioactive derivatives, collectively referred to as jasmonates (JAs), regulate a wide range of physiological processes in higher plants. JAs have a well-established role in orchestrating genome-wide transcriptional changes in response to biotic stress and developmental cues (1–4). In general, JAs promote defensive and reproductive processes, as well as inhibit the growth and photosynthetic output of vegetative tissues. These juxtaposing activities suggest a general role for the hormone in controlling resource allocation between growth- and defense-related processes, thus optimizing plant fitness in rapidly changing and hostile environments.
Recent studies indicate that JA controls the expression of early response genes by promoting the ubiquitin-dependent degradation of jasmonate ZIM domain (JAZ) proteins (5–7). The JAZ family of proteins in Arabidopsis consists of 12 members, which have been classified as a subgroup of the larger family of TIFY proteins that share a conserved TIF[F/Y]XG motif within the ZIM domain (8). A second defining feature of JAZs is the highly conserved Jas motif, which has a SLX2FX2KRX2RX5PY consensus sequence near the C terminus (5–7). Genetic analysis indicates that coronatine-insensitive 1 (COI1), an F-box protein that determines the target specificity of the E3 ubiquitin ligase SCFCOI1 (where SCF indicates Skp/Cullin/F-box), is required for most, if not all, JA-signaled processes (9–11). Arabidopsis JAZ3 (also known as JAI3) interacts with and presumably represses the activity of the MYC2 transcription factor that promotes the expression of JA-responsive genes (6). Current models indicate that degradation of JAZ repressors by the SCFCOI1–ubiquitin–proteasome pathway in response to a bioactive JA signal relieves the inhibition of MYC2, thereby activating the expression of early response genes. Functional analysis of the tomato homologs of COI1, JAZ, and MYC2 (5, 11, 12) indicates that this general mechanism of JA signaling is conserved in the plant kingdom.
The JAR1 gene encodes a JA–amido synthetase that catalyzes the formation of jasmonoyl-l-isoleucine (JA–Ile) (13–15). The JA-insensitive phenotype of jar1 mutant plants indicates that JA–Ile is an active signal in the JA pathway. Analysis of jar mutants also showed that JA–Ile is important for the regulation of plant defense responses to attack by pathogens and insects (14, 16, 17). Thines et al. (5) recently showed that formation of both Arabidopsis and tomato COI1–JAZ1 complexes is stimulated by JA–Ile but not by jasmonic acid, methyl-JA (MeJA), or the JA precursor 12-oxo-phytodienoic acid (OPDA). Endogenous JA–Ile levels increase within minutes of tissue damage, coincident with the expression of early JA-response genes (18). It is not known whether the JA–Ile-dependent interaction with COI1 is unique to JAZ1 or is more generally applicable to other members of the JAZ family.
The mechanism by which JA–Ile promotes COI1 binding to JAZ proteins remains to be determined. In yeast and animal cells, target recognition by E3 ubiquitin ligases typically depends on phosphorylation or other posttranslational modifications of the substrate (19). Notably, COI1 is homologous to TIR1, which functions as a receptor for the plant hormone auxin (20). Auxin regulates gene expression by binding to TIR1 and stimulating the ubiquitin–proteasome-dependent degradation of Aux/IAA transcriptional repressors (21, 22). Despite the many similarities between auxin and JA signaling, the identity of the JA receptor and its physiological ligand(s) is not known.
Coronatine (COR) is a phytotoxin produced by some plant pathogenic strains of Pseudomonas syringae (23). Several lines of evidence indicate that COR exerts its virulence effects by activating the host's JA signaling pathway (9, 24–27). The insensitivity of coi1 mutants of Arabidopsis and tomato to COR demonstrated that COI1 is required for the action of the toxin (9, 24). These observations, together with the structural similarity of COR to JA–Ile, support the notion that this virulence factor acts as a molecular mimic of JA–Ile (14, 28).
To test directly the hypothesis that JA–Ile and COR share a common molecular mechanism of action, we used an in vitro pull-down assay to assess the ability of COR, JA–Ile, and structurally related JA conjugates to promote the interaction of tomato COI1 with two divergent tomato JAZ proteins. Here, we provide evidence that COI1, or possibly a COI1–JAZ complex, is a receptor for JA–Ile and that COR exerts its virulence effects by functioning as a potent agonist of this receptor system. We also show that the Jas motif-containing C-terminal region of JAZ3 is necessary and sufficient for hormone-induced interaction of COI1 with JAZ3.
Results
Evidence That JA–Ile Directly Promotes COI1–JAZ1 Interaction in Vitro.
We previously used an in vitro pull-down assay to demonstrate that JA–Ile stimulates interaction between tomato COI1 and a purified JAZ1–His fusion protein in the absence of intact cells (5). Because these experiments used supernatants of tomato leaf extracts, after low-speed centrifugation, as a source of epitope-tagged COI1 (i.e., COI1–Myc), a potential role for cell membranes or membrane-bound proteins in JA–Ile action could not be excluded. However, depletion of cell membranes from COI1–Myc-containing tomato leaf extracts by high-speed centrifugation did not affect the amount of COI1–Myc recovered by JAZ1–His [supporting information (SI) Fig. S1]. This finding indicates that JA–Ile action in this cell-free system is mediated by soluble components.
The potential involvement of phosphorylation in JA signaling (29, 30) led us to test whether COI1–JAZ1 binding is modulated by reversible protein phosphorylation. The addition of protein kinase (staurosporine) and phosphatase inhibitors to pull-down assays did not affect the recovery of COI1–Myc by JAZ1–His (Fig. S2). This result indicates that COI1–JAZ1 interaction in response to JA–Ile likely occurs independently of protein phosphorylation or dephosphorylation. To test further whether JA–Ile action depends on an enzymatic activity in 35S-COI1–Myc leaf extracts, we investigated the effect of temperature on JA–Ile-dependent recovery of COI1–Myc by JAZ1–His. Recovery of COI1–Myc in reactions incubated at 16 and 30°C was severely diminished in comparison with a reaction carried out at the standard assay temperature, 4°C (Fig. S3). This finding argues against the possibility that enzymatic activity is required for JA–Ile-mediated COI1–JAZ1 interaction. It is possible that loss of COI1–JAZ1 interaction at higher temperatures results from JA–Ile metabolism or instability of the receptor complex. Collectively, these results suggest that COI1–JAZ interaction does not require a JA–Ile-induced enzymatic modification of either protein but rather that JA–Ile directly promotes the protein–protein interaction.
Requirement of Jasmonoyl–Amino Acid Conjugates for COI1 Interaction with Two Divergent JAZs.
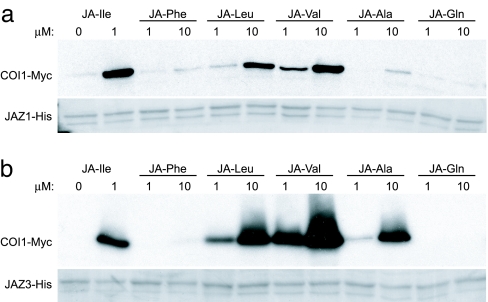
Recovery of COI1–Myc by JAZ1–His was not promoted by jasmonic acid (at concentrations up to 1 mM; data not shown), MeJA, or the JA precursor OPDA (5). Other plant hormones, including salicylic acid, abscisic acid, and indole-3-acetic acid, were also inactive (Fig. S4). To further define the specificity of JA–Ile as a signal for COI1–JAZ1 interaction, we compared the activity JA–Ile with other naturally occurring jasmonoyl–amino acid conjugates. JA–amino acid conjugates containing small hydrophobic amino acids stimulated COI1–JAZ1 binding to varying degrees, with the relative order of activity being JA–Ile → JA–Val → JA–Leu (Fig. 1a). JA–Ala was very weakly active, whereas JA–Phe and JA–Gln were inactive at the highest concentration tested (10 μM).
Fig. 1.
Specificity of JA–amino acid conjugates in promoting COI1–JAZ interaction. Pull-down assays were performed with recombinant JAZ1–His (a) or JAZ3–His (b) and extracts from 35S-COI1–Myc plants. Assays were supplemented with various JA–amino acid conjugates at the indicated concentration and incubated for 30 min at 4°C. Protein bound to JAZ1–His or JAZ3–His was analyzed by immunoblotting for the presence of COI1–Myc. The Coomassie Blue-stained blot in each panel shows the recovery of JAZ–His by the Ni affinity resin.
To determine whether the signal specificity of JAZ1 extends to other members of the tomato JAZ family, we identified a tomato JAZ cDNA whose sequence is significantly diverged from JAZ1. BLAST and phylogenetic analyses indicated that the tomato protein encoded by this cDNA is most similar to Arabidopsis JAZ3 (Fig. S5). We therefore refer to this tomato protein as SlJAZ3 (or hereafter simply as JAZ3). The sequence similarity between tomato JAZ1 and JAZ3 (≈27% amino acid identity) is restricted mainly to the ZIM domain and C-terminal Jas motif. Pull-down assays performed with a JAZ3–His fusion protein showed that the chemical specificity for COI1–Myc binding to JAZ3–His is very similar to that for JAZ1–His. For instance, the COI1–JAZ3 interaction was strongly promoted by JA–Ile, whereas no stimulatory effect was observed with jasmonic acid, MeJA, OPDA, JA–Phe, or JA–Gln (Fig. 1b; and data not shown). JA–Leu, –Val, and –Ala stimulated recovery of COI1–Myc by JAZ3–His to various extents, with the relative order of activity being JA–Ile ≈ JA–Val → JA–Leu → JA–Ala. JA–Val and JA–Ala were more effective in stimulating COI1–Myc binding to JAZ3–His than to JAZ1–His, suggesting that formation of COI1–JAZ1 complexes in vitro may be more selective for JA–Ile than for COI1–JAZ3 complexes. We conclude that recruitment of two divergent JAZ proteins by tomato COI1 is promoted specifically by JA–Ile and structurally related JA conjugates.
Coronatine Binds Directly to COI1–JAZ Complexes.
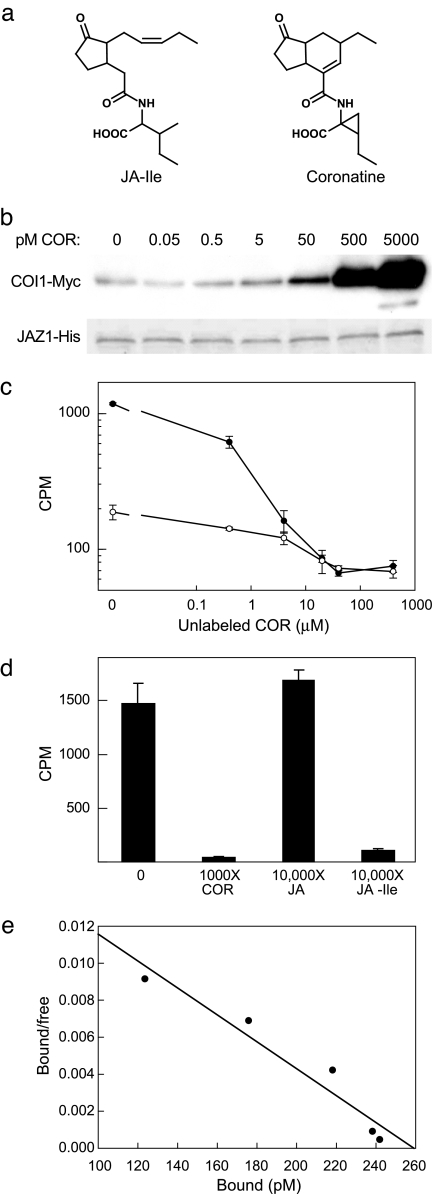
The ability of the P. syringae toxin COR to activate JA signaling in a COI1-dependent manner (9, 24, 26, 27), together with the structural similarities between COR and JA–Ile (Fig. 2a), led us to hypothesize that COR exerts its virulence effects by promoting COI1–JAZ interactions. We found that COR promotes COI1–Myc binding to JAZ1–His in a dose-dependent manner and, remarkably, that this stimulatory effect was apparent at concentrations of COR as low as 50 pM (Fig. 2b). Direct comparison of the activity of COR with that of JA–Ile showed that the toxin is ≈1,000-fold more active than JA–Ile (Fig. S6), which is in agreement with the previous observation that 50–500 nM concentrations of JA–Ile are required to stimulate COI1–JAZ1 interaction in this assay (5). COR was at least 100-fold more effective than JA–Ile in promoting the COI1–JAZ3 interaction in the JAZ3–His pull-down assay (data not shown).
Fig. 2.
Coronatine promotes formation of COI1–JAZ complexes. (a) Molecular structures of JA–Ile and COR. (b) JAZ1–His-containing pull-down assays supplemented with buffer (indicated by 0) or various concentrations of COR were processed as described in the legend to Fig. 1. The Coomassie blue-stained blot shows recovery of JAZ1–His. (c) Pull-down reactions containing extracts from 35S-COI1–Myc plants and JAZ3–His (filled circles) or JAZ1–His (open circles) were incubated with [3H]COR in the presence of increasing concentrations of unlabeled COR. Radioactivity recovered with JAZ–His is indicated (CPM). (d) Pull-down reactions containing 35S-COI1–Myc leaf extract, JAZ3–His, and [3H]COR were incubated in the absence (indicated by 0) or presence of the indicated amount of unlabeled COR, JA (jasmonic acid), or JA–Ile. (e) Pull-down reactions containing 35S-COI1–Myc leaf extract, JAZ3–His, and increasing concentrations of [3H]COR were used to construct a saturation curve for specific binding. Shown is a Scatchard plot of the saturation-binding data for a representative experiment. Error bars in c and d denote the SD of triplicate assays.
To determine whether COR is a ligand that directly binds to the COI1–JAZ complex, we performed binding assays in which crude leaf extracts containing COI1–Myc were incubated with [3H]COR and either JAZ1–His or JAZ3–His. The results in Fig. 2c show that both JAZ1–His and JAZ3–His retain [3H]COR after isolation of the fusion protein by Ni affinity chromatography and subsequent washing steps. The ability of unlabeled COR to compete with [3H]COR for binding indicates that the binding is specific. Binding of [3H]COR to JAZ3–His complexes was competed by the addition of 10,000-fold excess unlabeled JA–Ile but not by the addition of the same amount of jasmonic acid (Fig. 2d), indicating that the COR receptor eluting with JAZ3–His also binds to JA–Ile. Similar results were obtained in pull-down assays performed with JAZ1–His (Fig. S7). Scatchard analysis of saturation binding data obtained with the JAZ3–His pull-down assay showed that the apparent dissociation constant (Kd) of the receptor for COR was ≈20 nM (Fig. 2e).
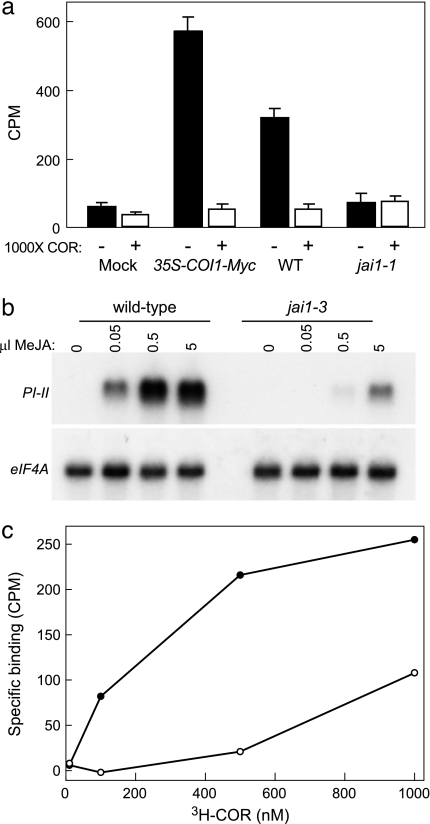
Specific binding of COR to the JAZ3–His complex was observed in pull-down reactions supplemented with crude leaf extract from WT tomato leaves (Fig. 3a). Thus, like COI1–Myc, endogenous COI1 interacts with JAZ3–His in a COR-dependent manner. To determine whether COI1 is required for COR binding, pull-down reactions were performed with leaf extracts from the COI1-deficient jai1-1 mutant of tomato that harbors a deletion in the SlCOI1 gene (11). As shown in Fig. 3a, jai1-1 extracts failed to promote recovery of [3H]COR by JAZ3–His. Binding assays performed with JAZ3–His in the absence of tomato leaf extract showed that COR does not bind specifically to JAZ3–His (Fig. 3a). We thus conclude that COI1 is an essential component of the COR/JA–Ile receptor and that JAZ protein alone is not sufficient for high-affinity ligand binding.
Fig. 3.
COI1 is an essential component of the jasmonate receptor. (a) Pull-down reactions containing JAZ3–His, [3H]COR, and crude leaf extract from the indicated tomato genotype (or an equivalent volume of buffer; indicated by Mock) were incubated in the presence (+) or absence (−) of 1,000-fold unlabeled COR. The amount of radioactivity recovered in the JAZ3–His complex is shown. Error bars denote the SD of triplicate assays. (b) Northern blot analysis of proteinase inhibitor II transcript accumulation in wild-type and jai1-3 plants treated in a closed container for 10 h with various amounts of vaporous MeJA. Blots were hybridized to an eIF4A cDNA as a loading control. (c) Specific binding of [3H]COR in pull-down assays containing JAZ3–His and leaf extract from either WT (closed circles) or jai1-3 (open circles) plants.
Homology between COI1 and the TIR1 auxin receptor provides indirect evidence for the idea that COI1 is a JA receptor (20, 31). To define further the role of COI1 in COR binding, we used the jai1-3 tomato mutant that harbors a point mutation (L418F) in the leucine-rich repeat domain of COI1 and, as a consequence, is partially insensitive to JA (Fig. S8). Northern blot analysis of the JA-inducible proteinase inhibitor II gene showed that jai1-3 leaves are ≈100-fold less responsive than WT leaves to exogenous MeJA (Fig. 3b). The mutated Leu residue in jai1-3 aligns with an Ile residue (Ile-406) in TIR1 that contacts the Aux/IAA peptide substrate within the auxin-binding pocket of TIR1 (20) (Fig. S9). We hypothesized that jai1-3 might impair COI1 interaction with either the ligand or the JAZ substrate. As shown in Fig. 3c, [3H]COR was recovered by JAZ3–His in pull-down assays supplemented with jai1-3 leaf extract but to a level that was much less than that obtained with WT extract. At 500 nM COR, for example, the recovery of specific binding in the presence of jai1-3 extract was only 1.3-fold above background, which was 10-fold lower than the amount of specific binding recovered in the presence of WT extract. These results indicate that the reduced sensitivity of jai1-3 leaves to exogenous JA correlates with reduced binding activity of jai1-3 extract to COR and provide evidence that Leu-418 of COI1 plays a role in the formation of a stable complex between COI1, JAZ, and COR.
The C-Terminal Region of JAZ3 Interacts with COI1 and Promotes Ligand Binding.
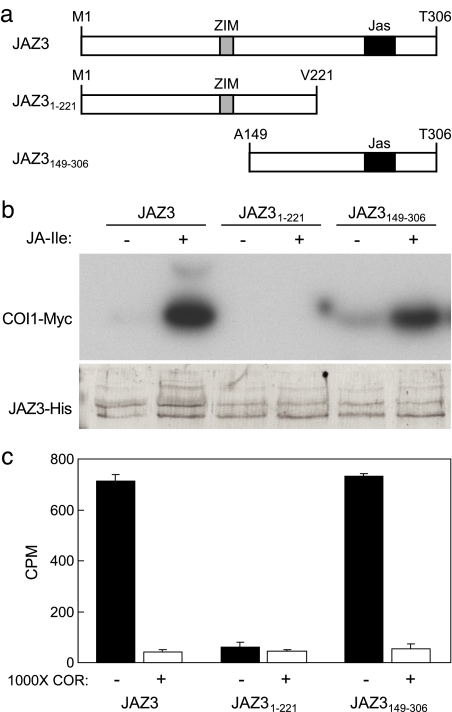
Having established that COR and JA–Ile bind to, and stimulate the formation of, a COI1–JAZ3 protein complex, we sought to identify the region of JAZ3 that facilitates this interaction. Truncated derivatives (JAZ31–221 and JAZ3149–306) of JAZ3 containing either the conserved TIFYXG motif in the ZIM domain or the Jas motif (Fig. 4a) were expressed as maltose-binding protein (MBP)–JAZ–His fusion proteins and tested for their ability to interact with COI1–Myc (Fig. 4a; Fig. S10). JAZ3149–306–His efficiently recovered COI1–Myc in a JA–Ile-dependent manner, whereas JAZ31–221–His did not (Fig. 4b). Moreover, we found that specific binding of [3H]COR was recovered by JAZ3149–306–His but not JAZ31–221–His in assays containing 35S-COI1–Myc extract (Fig. 4c). These results show that tomato JAZ3149–306 is necessary and sufficient for the COI1–JAZ3 interaction and specific binding of COR to the COI1–JAZ3 complex.
Fig. 4.
The C-terminal region of JAZ3 is required for COI1 interaction and specific binding of COR to the COI1–JAZ3 complex. (a) Schematic diagram of full-length and truncated JAZ3 constructs. The positions of the conserved ZIM (gray box) and Jas (black box) motifs are shown. The N-terminal MBP and C-terminal His6 fusions are not shown. (b) Pull-down assays containing 35S-COI1–Myc leaf extract and the indicated JAZ3 fusion protein were incubated in the presence (+) or absence (−) of 1 μM JA–Ile. Reactions were processed as described in the legend to Fig. 1. (c) Pull-down reactions containing 35S-COI1–Myc leaf extract, [3H]COR, and the indicated JAZ3 fusion protein were incubated in the presence (+) or absence (−) of unlabeled COR. The amount of radioactivity recovered in the JAZ3–His complex is shown.
Discussion
In this present study, we used a direct ligand-binding assay to investigate the mechanism by which COR and JA–Ile mediate the interaction between tomato COI1 and JAZ proteins. Our results indicate that JA–Ile does not act indirectly to induce an enzymatic modification of COI1 or JAZ but rather works directly to promote the COI1–JAZ interaction. The ability of JA–Ile to compete with COR for specific binding indicates that COR and JA–Ile are recognized by the same receptor. Because COR binding to the COI1–JAZ complex depends on COI1, we conclude that COI1 is an essential component of the perception apparatus. Moreover, that COR does not bind specifically to purified JAZ–His alone or in the presence of COI1-deficient tomato extract excludes the possibility that JAZ proteins function as JA receptors. These observations, together with the fact that JA–Ile-induced interaction between COI1 and JAZ does not require any other plant protein (5), provide strong evidence that COI1 is a receptor that specifically binds JA–Ile and COR.
Our results also provide evidence that the molecular mechanism of JA–Ile action is similar to that of auxin, which promotes substrate recruitment by creating a surface on the leucine-rich repeat domain of TIR1 that facilitates Aux–IAA binding (20). Detection of a stable ternary COI1–ligand–JAZ complex in the pull-down assay is consistent with the idea that JA–Ile/COR interact simultaneously with COI1 and JAZ. Because our binding assay relies on the recovery of components that copurify with JAZ–His, it remains to be determined whether COI1 binds to JA–Ile (or COR) in the absence of JAZ. A role for COI1 and JAZ as coreceptors thus remains a formal possibility. Significantly, however, COI1 is predicted to adopt a structure that is similar to the auxin receptor TIR1 (20). Several amino acid residues that mediate the interaction of TIR1 with auxin, substrate, and inositol hexakisphosphate are conserved in COI1. The results of binding experiments performed with extracts from the tomato jai1-3 mutant, which harbors a point mutation (L418F) in a region of COI1 that is homologous to the hormone-binding pocket of TIR1, suggests that this region of COI1 is important for interaction with the ligand and/or JAZ substrate. However, we cannot exclude the possibility that reduced binding of COR to jai1-3 extracts results from an effect of the L418F mutation on the stability or abundance of COI1.
We found that the interaction of COI1 with two divergent members (JAZ1 and JAZ3) of the tomato JAZ family is promoted in a highly specific manner by JA–Ile and structurally related JA conjugates. It is noteworthy that JA–Val, whose synthesis is catalyzed by JAR1-like enzymes (14, 32), was as active as JA–Ile in stimulating COI1 binding to JAZ3. This finding indicates that JA–Val may function as an endogenous signal for COI1-dependent responses, particularly in tissues that contain high JA–Val levels. JA–Leu and JA–Ala also exhibited activity, suggesting that these derivatives are bioactive JAs as well. Although several studies have suggested that jasmonic acid, MeJA, and OPDA are active per se as signals in the JA pathway (32–36), these compounds showed no activity in the JAZ1 and JAZ3 pull-down assays. It is possible that these nonconjugated derivatives promote COI1 interaction with JAZ proteins whose ligand specificity is different from that described here for JAZ1 and JAZ3. The well-defined repertoire of JAZ proteins in model plants such as Arabidopsis indicates that it will be possible to systematically determine the signal specificity of all JAZs by using protein–protein interaction assays.
Several pathovars of P. syringae possess a cluster of genes that direct the synthesis of the phytotoxin COR (23). Here, we show that COR functions as an agonist of the JA receptor. Remarkably, COR is ≈1000-fold more active than JA–Ile in promoting the COI1–JAZ3 interaction in vitro. This is consistent with the fact that COR is generally a much more potent signal in physiological responses than is jasmonic acid or MeJA (26, 37). The virulence properties of COR can thus be attributed to its ability to efficiently promote SCFCOI1-mediated ubiquitination and destruction of JAZ proteins. This conclusion is supported by studies demonstrating that COI1-deficient mutants of Arabidopsis and tomato are much less susceptible to infection by COR-producing strains of P. syringae (9, 24). Targeting of eukaryotic hormone receptors by pathogen virulence factors would appear to provide an efficient mechanism to manipulate genome-wide transcriptional programs and other processes that effectively suppress host cell defenses. It is not yet clear whether the toxic properties of COR result solely from an overstimulation of the JA signaling pathway due to its high receptor-binding efficiency or whether COR has additional properties that alter normal cellular processes.
Pull-down experiments with truncated forms of tomato JAZ3 showed that JAZ3149–306 interacts with COI1 in a ligand-dependent manner, whereas a derivative consisting of the N-terminal 221 aa does not. This finding indicates that the sequence determinants for substrate recognition by SCFCOI1 are located within the C-terminal 157 aa of JAZ3. Because the highly conserved Jas motif is located in this region, we suggest that the Jas motif mediates JAZ binding to COI1. This hypothesis is consistent with studies showing that deletion of the Jas motif stabilizes JAZ proteins against COI1-dependent degradation during JA signaling (5–7). Experiments conducted with Arabidopsis JAZ3, however, showed that the ZIM domain-containing N-terminal region, but not the C-terminal region containing the Jas motif, interacts with COI1 in the absence of exogenous JA (6). One possible explanation for these apparently disparate results is that COI1 interacts with the N and C termini of JAZ3 in a JA-independent and -dependent manner, respectively. Although we did not detect COI1–JAZ31–221 interaction in the absence of JA–Ile, our pull-down assay is likely less sensitive than the assay used by Chini et al. (6), who used radiolabeled proteins for their binding studies.
In summary, our results build on recent studies (5–7, 18) to define a unifying model for JA signaling in which direct recognition of JA–Ile by COI1 is coupled to ubiquitin-mediated degradation of JAZs and subsequent derepression of primary response genes. The results extend the emerging paradigm (20–22) of F-box proteins as intracellular sensors of small molecules to trigger the destruction of regulatory proteins and suggest a common evolutionary origin of the JA and auxin response pathways.
Materials and Methods
Biological Materials.
Growth conditions for Solanum lycopersicum (tomato) were described previously (38). A 35S-COI1–Myc transgenic line of tomato (cv Microtom) was used as a source of COI1–Myc in pull-down experiments (5). The tomato jai1-1 and jai1-3 (previously called spr5) mutants (cv Castlemart) were isolated and propagated as described in refs. 11 and 38 and in Fig. S8.
Chemicals.
Coronatine (C8115), phosphatase inhibitor mixture 1 (P2850), staurosporine (S4400), indole-3-acetic acid (I-2886), salicylic acid (S7401), and (±)-JA were purchased from Sigma. Jasmonoyl–amino acid conjugates were prepared, and the structures were verified by GC-MS as described previously (14, 39). The naturally occurring (−)-JA–Ile isomer was separated from (+)-JA–Ile by HPLC. (−)-JA–Ile was used in pull-down assays, whereas all other JA conjugates were used as a mixture of the (+)-JA– and (−)-JA–amino acid diastereomers. [3H]COR was prepared commercially (GE Healthcare) by using hydrogen–tritium exchange between tritiated water and unlabeled COR. The specific activity of [3H]COR as determined by mass spectrometry is 333 GBq per mmol. The radiochemical purity was 97.0% as determined by HPLC.
Cloning and Expression of JAZ Fusion Proteins.
A full-length cDNA for SlJAZ3 was PCR-amplified from a tomato EST clone (EST555543; BI935654) provided by the SOL Genomics Network (Cornell University). Primers used for the PCR were 5′-GCGCGGCCGCCGAGATGGAGAGGGACTTTATGG-3′ and 5′-ACGCGTCGACTAGCTTGGTCTCCTTACCG-3′. The resulting PCR product was cleaved with NotI and SalI and cloned into the corresponding restriction sites of pRMG-nMAL (5) to make the MBP–JAZ3–His6 fusion plasmid. Truncated derivatives of JAZ3, designated JAZ31–221 and JAZ3149–306, were amplified by using the following two primer sets, respectively: 5′-GCGCGGCCGCCGAGATGGAGAGGGACTTTATGG-3′ and 5′-CCCTCGAGCACATTAGGTGGAGCCA-3′; 5′-GCGCGGCCGCCTGTGCTCCACCTAAT-3′ and 5′-CCTCGAGGGTCTCCTTACCGGCTAA-3′. The PCR products were cleaved with NotI and XhoI and cloned into the corresponding sites of pRMG-nMAL. Full-length JAZ3 and JAZ1 fusion proteins, as well as the JAZ31–221 and JAZ3149–306 truncations, were expressed in Escherichia coli and purified by Ni affinity chromatography (Fig. S10) as described (5).
Pull-Down and [3H]COR Binding Assays.
A 35S-COI1–Myc transgenic line of tomato was used as a source of COI1–Myc in a JAZ–His pull-down assay described previously (5). Unless otherwise indicated, the quantity of JAZ–His added to each reaction was 25 μg. Protein concentrations were determined with a BCA protein assay kit (Pierce). Standard [3H]COR-binding assays contained 50 μg of purified JAZ–His protein, 4 mg of total leaf protein from 35S::COI1–Myc plants or an otherwise indicated genotype, and 400 nM [3H]COR (1.86 μCi) in a final volume of 0.5 ml of binding buffer [50 mM Tris, pH 6.8/10% glycerol, 100 mM NaCl, 25 mM imidazole, 20 mM 2-mercapto-ethanol, 10 μM MG132, 0.1% Tween 20, and Complete Mini protease inhibitor tablet-EDTA free (Roche)] and were performed in triplicate. Reactions were incubated at 4°C for 30 min, after which 80 μl of Ni resin was added. After an additional 15-min incubation at 4°C, JAZ–His-bound Ni resin was washed three times on microcentrifuge spin columns with 0.25 ml of binding buffer at 4°C. JAZ–His was eluted from the resin with 100 μl of 300 mM imidazole. Radioactivity in the resulting eluent was measured by scintillation counting (MicroBeta Trilux; PerkinElmer) after the addition of 1 ml of scintillation fluid (Optiphase Supermix; PerkinElmer).
Saturation-binding experiments were performed with increasing concentrations of [3H]COR in the presence or absence of 100-fold excess unlabeled COR. For experiments by using jai1-1 extracts, pull-down reactions contained 5 mg of total leaf protein, 50 μg of JAZ3–His, and 400 nM [3H]COR with or without the addition of 400 μM unlabeled COR. Saturation-binding experiments comparing WT and jai1-3 leaf extracts were conducted by incubating 5 mg of leaf protein and 50 μg of JAZ3–His with increasing concentrations of [3H]COR (10, 100, 500, and 1,000 nM) in the presence or absence of 100-fold excess unlabeled COR.
Analysis of Proteinase Inhibitor II Expression.
Proteinase inhibitor II (PI-II) protein and mRNA levels were measured according to published procedures (11, 38). Probed RNA blots were visualized with a phosphor imaging device, and the signal intensities quantified with the Quantity One-4.2.2 program (Bio-Rad). Values for each time point were normalized to the eIF4A loading control.
Supplementary Material
Acknowledgments.
We thank Sastry Jayanty and Hoo Sun Chung for helpful assistance with this work. Tomato EST clones were obtained from the SOL Genomics Network at Cornell University. This work was supported by the National Institutes of Health (G.A.H. and S.Y.H.), the U.S. Department of Energy (G.A.H. and S.Y.H.), the Michigan Agricultural Experiment Station at Michigan State University (G.A.H. and S.Y.H.), and the University of Nebraska Agricultural Research Division, supported in part by funds from the Hatch Act (P.E.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The nucleotide sequence of SlJAZ3 has been deposited in the GenBank database (accession no. EU194561).
This article contains supporting information online at www.pnas.org/cgi/content/full/0802332105/DCSupplemental.
References
- 1.Balbi V, Devoto A. Jasmonate signalling network in Arabidopsis thaliana: Crucial regulatory nodes and new physiological scenarios. New Phytol. 2008;177:301–318. doi: 10.1111/j.1469-8137.2007.02292.x. [DOI] [PubMed] [Google Scholar]
- 2.Howe G, Jander G. Plant immunity to insect herbivores. Annu Rev Plant Biol. 2008;59:41–66. doi: 10.1146/annurev.arplant.59.032607.092825. [DOI] [PubMed] [Google Scholar]
- 3.Browse J, Howe GA. New weapons and a rapid response against insect attack. Plant Physiol. 2008;146:832–838. doi: 10.1104/pp.107.115683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wasternack C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot (London) 2007;100:681–697. doi: 10.1093/aob/mcm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thines B, et al. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 6.Chini A, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 7.Yan Y, et al. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell. 2007;19:2470–2483. doi: 10.1105/tpc.107.050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vanholme B, Grunewald W, Bateman A, Kohchi T, Gheysen G. The tify family previously known as ZIM. Trends Plants Sci. 2007;12:239–244. doi: 10.1016/j.tplants.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Feys B, Benedetti CE, Penfold CN, Turner JG. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell. 1994;6:751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- 11.Li L, et al. The tomato homolog of Coronatine-Insensitive 1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell. 2004;16:126–143. doi: 10.1105/tpc.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boter M, Ruiz-Rivero O, Abdeen A, Prat S. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 2004;18:1577–1591. doi: 10.1101/gad.297704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Staswick PE, Tiryaki I, Rowe ML. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell. 2002;14:1405–1415. doi: 10.1105/tpc.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Staswick PE, Tiryaki I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell. 2004;16:2117–227. doi: 10.1105/tpc.104.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suza WP, Staswick PE. The role of JAR1 in jasmonoyl-l-isoleucine production in Arabidopsis wound response. Planta. 2008;227:1221–1232. doi: 10.1007/s00425-008-0694-4. [DOI] [PubMed] [Google Scholar]
- 16.Staswick PE, Yuen GY, Lehman CC. Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J. 1998;15:747–754. doi: 10.1046/j.1365-313x.1998.00265.x. [DOI] [PubMed] [Google Scholar]
- 17.Kang JH, Wang L, Giri A, Baldwin IT. Silencing threonine deaminase and JAR4 in Nicotiana attenuata impairs jasmonic acid–isoleucine-mediated defenses against Manduca sexta. Plant Cell. 2006;18:3303–3320. doi: 10.1105/tpc.106.041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung HS, et al. Regulation and function of Arabidopsis JAZ genes in response to wounding and herbivory. Plant Physiol. 2008;146:952–964. doi: 10.1104/pp.107.115691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deshaies RJ. SCF and cullin/RING H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 20.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 21.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 22.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 23.Bender CL, Alarcon-Chaidez F, Gross DC. Pseudomonas syringae phytotoxins: Mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev. 1999;63:266–292. doi: 10.1128/mmbr.63.2.266-292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Y, et al. Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J. 2003;36:485–499. doi: 10.1046/j.1365-313x.2003.01895.x. [DOI] [PubMed] [Google Scholar]
- 25.Lauchli R, Boland W. Indanoyl amino acid conjugates: Tunable elicitors of plant secondary metabolism. Chem Rec. 2003;3:12–21. doi: 10.1002/tcr.10043. [DOI] [PubMed] [Google Scholar]
- 26.Uppalapati SR, et al. The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. Plant J. 2005;42:201–217. doi: 10.1111/j.1365-313X.2005.02366.x. [DOI] [PubMed] [Google Scholar]
- 27.Thilmony R, Underwood W, He SY. Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J. 2006;46:34–53. doi: 10.1111/j.1365-313X.2006.02725.x. [DOI] [PubMed] [Google Scholar]
- 28.Krumm T, Bandemer K, Boland W. Induction of volatile biosynthesis in the lima bean (Phaseolus lunatus) by leucine- and isoleucine conjugates of 1-oxo- and 1-hydroxyindan-4-carboxylic acid: Evidence for amino acid conjugates of jasmonic acid as intermediates in the octadecanoid signalling pathway. FEBS Lett. 1995;377:523–529. doi: 10.1016/0014-5793(95)01398-9. [DOI] [PubMed] [Google Scholar]
- 29.Rojo E, et al. Reversible protein phosphorylation regulates jasmonic acid-dependent and -independent wound signal transduction pathways in Arabidopsis thaliana. Plant J. 1998;13:153–165. doi: 10.1046/j.1365-313x.1998.00020.x. [DOI] [PubMed] [Google Scholar]
- 30.Katou S, et al. Involvement of PPS3 phosphorylated by elicitor-responsive mitogen-activated protein kinases in the regulation of plant cell death. Plant Physiol. 2005;139:1914–1926. doi: 10.1104/pp.105.066795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parry G, Estelle M. Auxin receptors: A new role for F-box proteins. Curr Opin Cell Biol. 2006;18:152–156. doi: 10.1016/j.ceb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Wang L, Allmann S, Wu J, Baldwin IT. Comparisons of LOX3- and JAR4/6-silenced plants reveal that JA and JA–AA conjugates play different roles in herbivore resistance of Nicotiana attenuata. Plant Physiol. 2008;146:904–915. doi: 10.1104/pp.107.109264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stintzi A, Weber H, Reymond P, Browse J, Farmer EE. Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc Natl Acad Sci USA. 2001;98:12837–12842. doi: 10.1073/pnas.211311098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seo HS, et al. Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc Natl Acad Sci USA. 2001;98:4788–4793. doi: 10.1073/pnas.081557298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kramell R, et al. Synthesis of N-(jasmonoyl)amino acid conjugates. FEBS Lett. 1997;414:197–202. doi: 10.1016/s0014-5793(97)01005-3. [DOI] [PubMed] [Google Scholar]
- 36.Koch T, Krumm T, Jung V, Engelberth J, Boland W. Differential induction of plant volatile biosynthesis in the lima bean by early and late intermediates of the octadecanoid-signaling pathway. Plant Physiol. 1999;121:153–162. doi: 10.1104/pp.121.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koda Y, et al. Similarities of the biological activities of coronatine and coronafacic acid to those of jasmonic acid. Phytochemistry. 1996;41:93–96. .38. [Google Scholar]
- 38.Li L, Howe GA. Alternative splicing of prosystemin pre-mRNA produces two isoforms that are active as signals in the wound response pathway. Plant Mol Biol. 2001;46:409–419. doi: 10.1023/a:1010645330275. [DOI] [PubMed] [Google Scholar]
- 39.Kramell R, Schmidt J, Schneider G, Sembdner G, Schreiber K. Amino acid conjugates of jasmonic acid induce jasmonate-responsive gene expression in barley (Hordeum vulgare L.) leaves. Tetrahedron. 1988;44:5791–5807. doi: 10.1016/s0014-5793(97)01005-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.