Abstract
Glucose is a fundamental metabolite, yet how cells sense and respond to changes in extracellular glucose concentration is not completely understood. We recently reported that the MondoA:Mlx dimeric transcription factor directly regulates glycolysis. In this article, we consider whether MondoA:Mlx complexes have a broader role in sensing and responding to glucose status. In their latent state, MondoA:Mlx complexes localize to the outer mitochondrial membrane, yet shuttle between the mitochondria and the nucleus. We show that MondoA:Mlx complexes accumulate in the nucleus in response to glucose and 2-deoxyglucose (2-DG). Furthermore, nuclear localization of MondoA:Mlx depends on the enzymatic activity of hexokinases. These enzymes catalyze conversion of glucose to glucose-6-phosphate (G6P), which is the first step in the glycolytic pathway. Together, these findings suggest that MondoA:Mlx monitors intracellular G6P concentration and translocates to the nucleus when levels of this key metabolite increase. Transcriptional profiling experiments demonstrate that MondoA is required for >75% of the 2-DG-induced transcription signature. We identify thioredoxin-interacting protein (TXNIP) as a direct and glucose-regulated MondoA:Mlx transcriptional target. Furthermore, MondoA:Mlx complexes, via their regulation of TXNIP, are potent negative regulators of glucose uptake. These studies suggest a key role for MondoA:Mlx complexes in the adaptive transcriptional response to changes in extracellular glucose concentration and peripheral glucose uptake.
Keywords: metabolism, mitochondria, transcription
Glucose is an abundant and universal nutrient for prokaryotic and eukaryotic cells, providing carbons for biosynthetic reactions and ATP for energy. In mammals, misregulation of circulating glucose levels, glucose uptake, and intracellular glucose homeostasis underlies metabolic disorders such as diabetes (1). Furthermore, an elevated glycolytic rate is a near universal feature of tumor cells (2). These two points underscore the absolute necessity for cells to tightly control how they sense and respond to changes in intracellular glucose concentration, yet we know relatively little about the precise molecular mechanisms by which mammalian cells sense and respond transcriptionally to glucose (1, 3).
We recently demonstrated that MondoA:Mlx heterodimers transcriptionally activate a number of important glycolytic target genes including hexokinase II (HKII), 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (PFKFB3), and lactate dehydrogenase A (LDH-A) (4). MondoA and Mlx are members of the basic helix–loop–helix-leucine zipper (bHLHZip) family. A dominating feature of MondoA:Mlx complexes is their localization to the outer mitochondrial membrane (OMM) (4). When artificially targeted to the nucleus, MondoA:Mlx complexes bind to CACGTG E-box sequences and appear to have a primary role in transcription activation (4, 5). These findings suggest the nuclear activity of MondoA:Mlx complexes is under tight regulatory control dictated by regulation of their subcellular localization (5, 6). Indeed, MondoA:Mlx complexes shuttle between the OMM and nucleus, suggesting that they facilitate communication between these two essential organelles (4). Whether MondoA:Mlx complexes regulate gene expression through this “shuttling” activity or whether there is a “trigger” for nuclear localization is unknown.
Several lines of evidence hint at an important role for MondoA:Mlx complexes in regulating the cellular transcriptional response to glucose. First, given the essential role of mitochondria in bioenergetics, we proposed that MondoA:Mlx complexes sense changes in intracellular energy state at the OMM and accumulate in the nucleus to guide an adaptive transcriptional response (4). Second, our knowledge of genes regulated by MondoA:Mlx complexes is still somewhat limited; however, HKII, PFKFB3, and LDH-A are direct transcriptional targets (4). Consistent with their regulation of these glycolytic target genes, MondoA:Mlx complexes are necessary and sufficient regulators of glucose flux (4). Finally, Gal4-DNA binding domain tethering assays suggest that the transcription activation domain of MondoA requires glucose for full activity (7).
Here, we investigate how the subcellular localization and transcriptional activity of MondoA:Mlx complexes are affected by changes in glucose concentration. We show that MondoA:Mlx complexes contribute to the majority of glucose-induced transcription by accumulating in the nucleus in response to an elevated intracellular energy state likely by sensing glucose-6-phosphate (G6P) levels. Further, we show that MondoA:Mlx complexes are potent negative regulators of glucose uptake via the direct and glucose-dependent regulation of thioredoxin-interacting protein (TXNIP). Together, our data suggest a key role for MondoA:Mlx complexes in the adaptive transcriptional response required for proper glucose utilization.
Results
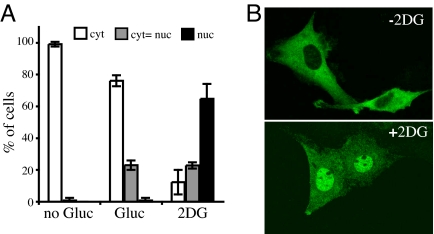
MondoA:Mlx complexes localize to the OMM, yet they are potent transcriptional activators, suggesting that they might accumulate in the nucleus in response to intracellular or extracellular signaling events. Because MondoA:Mlx complexes regulate glycolytic targets and glycolysis (4), we tested whether known inducers of glycolysis, such as hypoxia or increased AMP levels, resulted in the nuclear accumulation of MondoA. In rat L6 myoblasts transfected with MondoA and Mlx, MondoA remained in the cytoplasm after 8 h growth in 1% oxygen or treatment with AICAR, an AMP mimic that activates the AMP-activated protein kinase (AMPK) (data not shown). We next considered whether MondoA might function more directly in sensing glucose itself or glucose-derived metabolites. Consistent with this hypothesis, treatment with high glucose after glucose starvation resulted in an increase in cells expressing MondoA in the nucleus; however, the effect was not complete. Treatment with the glucose analog, 2-deoxyglucose (2-DG), resulted in a more pronounced nuclear localization of MondoA and its partner Mlx in the majority of cells (Fig. 1 and data not shown). Glucose and 2-DG are phosphorylated by hexokinases to generate glucose-6 phosphate (G6P) and 2-DG-6-phosphate (2-DG6P), respectively. G6P does not accumulate to high levels because it rapidly enters the glycolytic and pentose phosphate pathways. By contrast, 2-DG6P accumulates because it cannot be processed further by glycolysis and only enters the pentose phosphate pathway at a very reduced rate (8). As such, we suggest that under physiological signaling conditions MondoA senses G6P levels, accounting for the partial affect of glucose on MondoA nuclear localization. The supraphysiological accumulation of 2-DG6P likely accounts for the more dramatic affect of 2-DG.
Fig. 1.
MondoA is a glucose sensor. (A) Rat L6 cells expressing MondoA and Mlx were starved overnight and treated with glucose or 2-DG as indicated. Subcellular localization of MondoA was determined after 6 h of treatment. Localization of Mlx was similar (data not shown). (B) Representative images of MondoA subcellular localization in the absence or presence of 2-DG. (Magnification: ×40.)
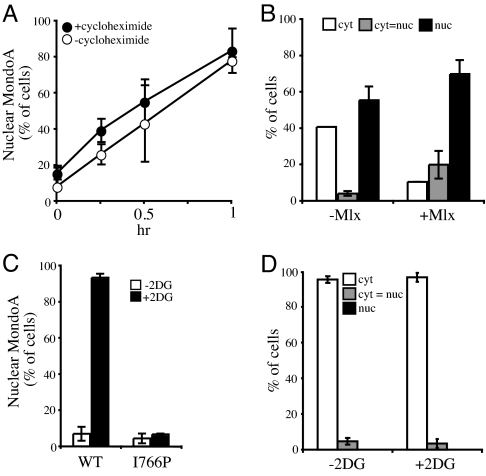
We next determined the time course of MondoA nuclear translocation in response to 2-DG treatment. Translocation of MondoA to the nucleus could be detected as early as 15 min after 2-DG treatment and continued to increase until 1 h posttreatment when ≈80% of the cells showed a nuclear localization of MondoA (Fig. 2A). The kinetics of MondoA nuclear translocation in the presence of the protein synthesis inhibitor cycloheximide were nearly identical to the untreated controls, suggesting that preexisting MondoA, rather than de novo synthesized protein, translocates to the nucleus as an immediate early response to 2-DG treatment.
Fig. 2.
Nuclear translocation and accumulation of MondoA is an immediate-early response to 2-DG and requires Mlx. (A) L6 cells were treated with 2-DG in the presence or absence of cycloheximide, and the amount of nuclear MondoA was determined at the indicated time points. (B) L6 cells expressing MondoA alone or MondoA and Mlx as indicated were treated with 2-DG, and the subcellular localization of MondoA was determined after 3 h. (C and D) L6 cells expressing WT MondoA or the dimerization-defective MondoA mutant, MondoA(I766P), in combination with Mlx were treated with 2-DG and the nuclear localization of MondoA or MondoA(I766P) (C) or Mlx in the presence of MondoA(I766P) (D), determined after 3 h.
Mlx is absolutely required for the nuclear translocation of MondoA (6). We performed two experiments to test whether MondoA requires interaction with Mlx to respond to 2-DG. First, in the absence of overexpressed Mlx, MondoA was more cytoplasmic after 2-DG treatment than in the presence of Mlx (Fig. 2B). Endogenous Mlx presumably mediates the partial 2-DG effect on MondoA translocation in the absence of transfected Mlx. Second, we used MondoA(I766P) with a mutation in the leucine zipper domain that abrogates Mlx binding to determine whether physical interaction between the two proteins is essential for nuclear accumulation. Both MondoA(I766P) and Mlx remained in the cytoplasm after 2-DG treatment (Fig. 2 C and D). Therefore, dimerization between MondoA and Mlx is essential for their response to 2-DG, suggesting that MondoA:Mlx complexes, rather than the protein monomers, are the regulated species.
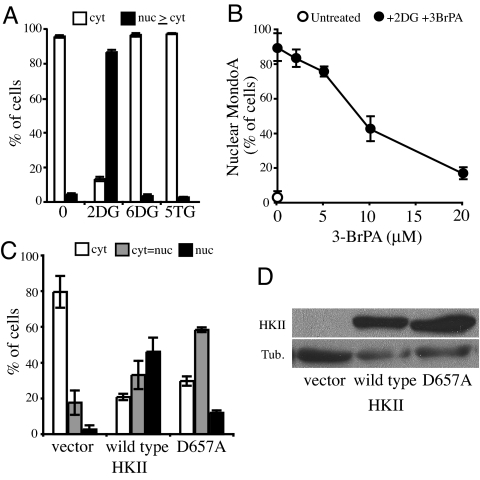
Our model that MondoA senses G6P suggests a critical role of hexokinases in controlling the nuclear accumulation of MondoA in response to 2-DG. We used several approaches to test this hypothesis. First, we determined the effect of two additional glucose analogs on MondoA translocation: 6-deoxyglucose (6-DG), which cannot be phosphorylated by hexokinases, or 5-thio-glucose (5-TG), which is a competitive inhibitor of hexokinases (9). In contrast to the effect of 2-DG, MondoA remained in the cytoplasm after treatment with either 6-DG or 5-TG (Fig. 3A). Second, we determined whether the hexokinase inhibitor 3-bromopyruvate (3-BrPA) altered the 2-DG response of MondoA (10). Treatment with 3-BrPA blocked 2-DG-dependent translocation of MondoA in a dose-dependent manner (Fig. 3B). 3-BrPA can also inhibit GAPDH (11), another glycolytic enzyme that functions downstream of hexokinases in the glycolytic pathway. 2-DG6P is not processed further by glycolysis and therefore cannot be metabolized to a substrate for GAPDH. As such, we propose that the inhibition of 2-DG-induced nuclear accumulation of MondoA by 3-BrPA is primarily via its inhibition of hexokinase rather than inhibition of GAPDH activity.
Fig. 3.
Nuclear translocation of MondoA depends on hexokinase activity. (A) HA1ER cells expressing MondoA and Mlx were treated with the indicated sugars, and subcellular localization of MondoA was determined after 3 h. (B) HA1ER cells expressing MondoA and Mlx were treated with 2-DG in the presence of the indicated increasing concentrations of 3-BrPA, and subcellular localization of MondoA was determined after 3 h. (C) HEK293T cells transfected with WT HKII or a catalytically impaired HKII mutant, HKII(D657A), were treated with 2-DG, and the subcellular localization of MondoA was determined after 3 h. (D) Expression of HKII and HKII (D657A) was identical in this experiment.
We have observed 2-DG-induced nuclear translocation of MondoA in L6 and C2C12 myoblasts, NIH 3T3 and BJ fibroblasts, K562 erythroblasts, and A549 and HA1E/HA1ER epithelial cells, suggesting that MondoA responds broadly and generally to 2-DG (data not shown). In contrast, in HEK293T cells, MondoA:Mlx failed to accumulate in the nucleus in response to 2-DG (Fig. 3C). HEK293T cells do not express HKII (Fig. 3D) suggesting that they may not convert 2-DG to 2-DG6P efficiently. We therefore introduced WT HKII into HEK293T cells and found that MondoA:Mlx now accumulated in the nucleus in response to 2-DG (Fig. 3C). By contrast, introduction of a HKII mutant (D657A) reported to be severely catalytically impaired (12) only weakly rescued the 2-DG-response defect of HEK293T cells (Fig. 3C). Together, these experiments suggests that the nuclear accumulation of MondoA:Mlx complexes in response to 2-DG treatment requires the catalytic activity of hexokinases.
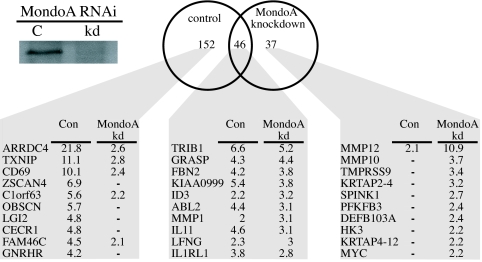
Our data suggest that MondoA:Mlx complexes sense elevated G6P levels and translocate to the nucleus to trigger an adaptive transcriptional response. To discover target genes regulated by MondoA:Mlx complexes under these conditions, we used expression profiling to identify 2-DG induced genes in control and MondoA knockdown HA1ER cells (Fig. 4). This analysis identified 198 targets induced by 2-DG >1.8-fold in the control cells [Fig. 4 and supporting information (SI) Fig. S1]. By contrast, only 83 genes were induced >1.8-fold in the MondoA knockdown cells. Importantly, 46 genes showed similar induction in both cell populations, indicating that both cell lines were capable of 2-DG-induced transcription and were not generally defective in glucose-induced transcription. Of the 198 2-DG-regulated genes, 152 targets were impaired in their 2-DG regulation in the MondoA knockdown cells. Thus, MondoA is required for ≈75% of 2-DG induced transcription in this cell line. More than 90% of the most highly 2-DG induced genes (those induced >3-fold) depended on MondoA, suggesting an even more prominent role for MondoA at highly regulated targets (data not shown). Finally, 37 targets were up-regulated by 2-DG only in the MondoA knockdown cells, suggesting cellular compensation to MondoA knockdown or a previously unappreciated transcriptional repressor function of MondoA:Mlx complexes.
Fig. 4.
MondoA is a master regulator of glucose-induced transcription. Genes induced >1.8-fold (P < 0.005) after a 3-h 2-DG treatment in control and MondoA knockdown cells were determined by using Agilent 44K human microarrays. The 10 most highly regulated genes in each class are indicated along with their fold up-regulation in the two cell populations. (Inset) MondoA levels in the control and knockdown cells used for this experiment.
To place the 2-DG-induced and MondoA-dependent target genes in a physiological context, we queried the Kyoto Encyclopedia of Genes and Genomes database. We identified 13 annotated pathways enriched with z-scores >2 in our target gene list (Fig. S2). The majority of these pathways, e.g., the Jak-STAT pathway, the MAPK pathway, and the TGF-β pathway, control cell proliferation and differentiation. These findings suggest additional coordination between growth control and nutrient-sensing pathways, similar to that described for the AMPK–mTOR pathway (13).
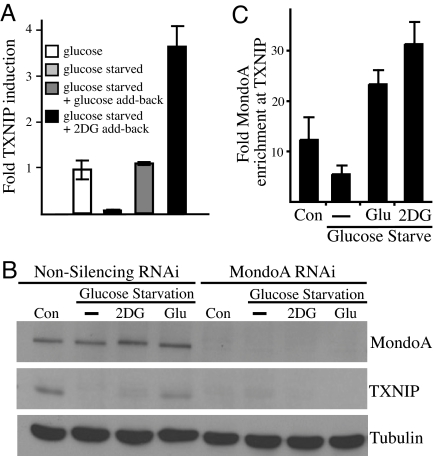
Arrestin domain-containing protein 4 (ARRDC4) was the most highly 2-DG induced and MondoA-dependent gene, followed by TXNIP. ARRDC4 and TXNIP are paralogs showing 63% similarity over their entire ORFs; however, there is little functional information available for ARRDC4 (14, 15). By contrast, TXNIP has multiple roles in cell physiology, including one in peripheral glucose disposal (16, 17). Further, TXNIP expression is regulated by glucose through two CACGAG carbohydrate response elements (ChoREs) in its promoter (18). The glucose-responsive transcription factor that activates TXNIP expression is not known; therefore, we evaluated the contribution of MondoA:Mlx by several approaches. First, TXNIP mRNA was down-regulated in glucose-starved HA1ER cells, but up-regulated by the addition of either glucose or 2-DG (Fig. 5A). 2-DG was more effective than glucose in inducing TXNIP mRNA expression, correlating well with the differential effects of 2-DG and glucose on MondoA:Mlx nuclear accumulation (Fig. 1A). To determine whether MondoA:Mlx complexes regulated TXNIP, we generated HA1ER cells expressing a nonspecific shRNA or expressing a MondoA specific shRNA (Fig. 5B). In control cells, TXNIP protein was down-regulated in the absence of glucose and was reexpressed by the addition glucose or 2-DG. Glucose induced TXNIP protein expression more effectively than 2-DG, presumably because translational efficiency was impaired in 2-DG-treated cells (13). By contrast, the constitutive expression and induction of TXNIP protein by glucose or 2-DG was completely blocked in MondoA knockdown cells. Therefore, MondoA is required for constitutive TXNIP expression and its induction after glucose or 2-DG addition.
Fig. 5.
MondoA is a direct and glucose-dependent regulator of TXNIP. (A) qRT-PCR was used to determine the relative expression of TXNIP in HA1ER cells treated as indicated. (B) Expression of TXNIP, MondoA, and tubulin was determined by Western blotting in control and MondoA knockdown HA1ER cells. Cells were treated as indicated. (C) ChIP was used to determine MondoA occupancy at the TXNIP promoter in HA1ER cells under the indicated conditions.
To determine whether the glucose-dependent regulation of TXNIP by MondoA:Mlx complexes is direct or indirect we performed ChIP. We immunoprecipitated MondoA from HA1ER cells grown in different media conditions and identified immunoprecipitated DNA by using primer pairs flanking the known ChoREs in the TXNIP promoter (18). MondoA:Mlx complexes occupied the TXNIP promoter under normal growth conditions, but occupancy was reduced after glucose depletion. Glucose and, to a greater extent, 2-DG addition restored binding of MondoA:Mlx complexes to the TXNIP promoter (Fig. 5C). These data suggest that MondoA:Mlx complexes are direct and glucose-dependent transcriptional regulators of TXNIP, activating expression after their glucose/2-DG-dependent nuclear accumulation and occupancy of the ChoREs of the TXNIP promoter.
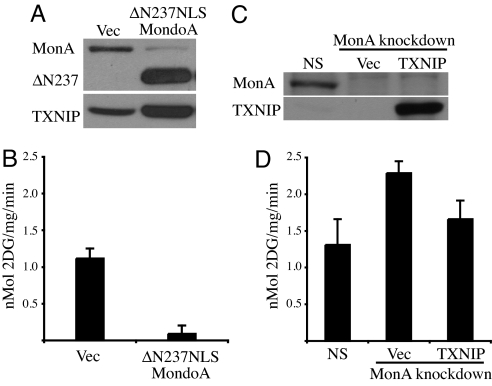
TXNIP has recently been implicated as an important negative regulator of glucose uptake in skeletal muscle (17). As such, we tested whether MondoA also controls glucose uptake via its regulation of TXNIP. Two experiments support this hypothesis. First, TXNIP protein was up-regulated in HA1E cells expressing a dominant active nuclear form of MondoA, ΔN237NLSMondoA, but not in mock-infected control cells (Fig. 6A). Glucose uptake was also dramatically down-regulated in the ΔN237NLSMondoA-expressing cells, consistent with TXNIP controlling glucose uptake downstream of MondoA:Mlx (Fig. 6B). We also observe a similar up-regulation of TXNIP and down-regulation of glucose uptake in rat L6 myoblasts demonstrating the generality of this finding (Fig. S3). Second, we determined rates of glucose uptake in control or MondoA knockdown HA1ER cells. MondoA knockdown cells, which have reduced TXNIP levels (Fig. 6C), exhibited an elevated rate of glucose uptake relative to control cells. Further, this elevation in glucose uptake was significantly reduced by overexpression of TXNIP (Fig. 6D). Together these two experiments demonstrate that MondoA is a potent negative regulator of glucose uptake, likely via its regulation of TXNIP.
Fig. 6.
MondoA is a negative regulator of glucose uptake. (A) TXNIP and MondoA expression in HA1E cells expressing ΔN237NLSMondoA (ΔN237), or vector alone, was determined by Western blotting. (B) Glucose uptake in control or ΔN237NLSMondoA expressing HA1E cells. (C) TXNIP and MondoA expression in control and MondoA knockdown HA1ER cells transfected with vector alone or a TXNIP expression vector was determined by Western blotting. (D) Glucose uptake in MondoA knockdown cells with or without overexpression of TXNIP.
Discussion
How mammalian cells sense and respond to changes in glucose concentration is not completely understood. Our data suggest a comprehensive model for how mammalian cells coordinate an adaptive transcriptional response to elevated extracellular glucose. Glucose sensing by MondoA:Mlx complexes in the cytoplasm and subsequent accumulation in the nucleus provides important insights into this essential physiological process. Our microarray experiments suggest that MondoA may contribute to upward of 75% of glucose-induced gene expression. To help validate our array findings, we examined the regulation of TXNIP in detail and show that its basal and glucose-induced expression exquisitely depends on MondoA. Finally, we establish MondoA as a potent negative regulator of glucose uptake via its regulation of TXNIP in vitro, suggesting an important role for MondoA in peripheral glucose disposal in vivo.
The first key aspect of our model of how MondoA:Mlx complexes sense and respond to glucose is their accumulation in the nucleus in response to 2-DG. We considered several possibilities to explain this finding. First, as 2-DG is a metabolic inhibitor, perhaps MondoA:Mlx complexes sense a nutrient depleted state. However, MondoA:Mlx complexes remained in the cytoplasm after glucose, amino acid, or ATP depletion (Fig. 1 and data not shown). Given these results, it seems unlikely that MondoA:Mlx sense a nutrient/energy depleted state. Second, 2-DG6P is not further processed by glycolysis, yet can enter the pentose phosphate shunt, but at a very reduced rate (8), suggesting that MondoA:Mlx complexes might sense a pentose phosphate shunt intermediate. However, MondoA:Mlx complexes still accumulated in the nucleus in response to 2-DG treatment after inhibition of G6P dehydrogenase, the first enzyme in the pentose phosphate shunt, by 6-amino-nicotinamide (data not shown). Therefore, we do not favor a model where MondoA:Mlx complexes sense the levels of a pentose phosphate shunt intermediate.
The final possibility arises from our observation that MondoA:Mlx complexes translocate to the nucleus partially in response to glucose and almost completely in response to 2-DG (Fig. 1A). Because translocation depends on the catalytic activity of hexokinases (Fig. 3), we propose that under physiological conditions, MondoA:Mlx complexes are extremely sensitive monitors of intracellular G6P concentration and translocate to the nucleus when G6P levels increase. G6P is quickly processed into the glycolytic and pentose phosphate pathways, which likely accounts for the partial response of MondoA:Mlx complexes to glucose (Fig. 1). By contrast, 2-DG6P is not readily metabolized, which explains the robust nuclear translocation of MondoA:Mlx complexes in the presence of 2-DG. The ability of MondoA:Mlx complexes to sense subtle changes in intracellular G6P concentrations, coupled with their very potent transcription activation domain (5), must allow for a robust transcriptional response to subtle changes in extracellular glucose concentration.
A second key feature of our model of glucose sensing by MondoA:Mlx complexes is the requirement of hexokinases for their nuclear accumulation. In yeast and plants, hexokinases are known to play roles in glucose sensing (19, 20). Our experiments suggest that they also contribute to glucose sensing in mammalian cells. For example, multiple experiments presented here suggest a critical role for hexokinase catalytic activity in the nuclear translocation of MondoA:Mlx complexes. Furthermore, like MondoA:Mlx complexes, two hexokinases (hexokinase I and II) localize to the OMM where they have preferential access to ATP synthesized in the mitochondria (9). The OMM localization of both hexokinase activity and MondoA:Mlx complexes suggests that their activities may be spatially coupled. Such a model would allow for a precise coordination of intracellular glucose and ATP concentrations, the substrates for hexokinases, with the transcriptional activity of MondoA:Mlx complexes. As hexokinases can be displaced from the OMM by their product G6P (9), a spatial coupling model suggests that an allosteric change in hexokinases may contribute to the nuclear accumulation of MondoA:Mlx complexes in response to glucose or 2-DG.
The third aspect of our model for the function of MondoA:Mlx complexes is their requirement for 2-DG-induced gene expression. Microarray analysis suggests that MondoA:Mlx complexes contribute to the majority of glucose/2-DG-induced transcription. Our work with TXNIP suggests that glucose-dependent occupancy and activation of its promoter by MondoA:Mlx complexes underlies its glucose-dependent transcriptional activation. We show that, at a minimum, MondoA:Mlx complexes are necessary for glucose-induced TXNIP expression; we cannot formally rule out requirement for other glucose-induced transcription factors or cofactors.
Our experiments show that MondoA is required for the majority of 2-DG-induced gene expression, yet many questions remain about how MondoA:Mlx complexes select and activate target genes. For example, our array experiments test only a proximal transcriptional effect of glucose sensing, because 2-DG6P is not further processed by glycolysis. As such, it will be of interest to determine whether MondoA can sense, and respond to, additional metabolites as glucose is processed further. In addition, we previously demonstrated that HKII, PFKFB3, and LDH-A are direct transcriptional targets of MondoA:Mlx complexes (4), yet none of these genes are strongly glucose/2-DG-regulated in HA1ER cells nor are they significantly down-regulated in MondoA knockdown cells (data not shown). At present, we do not know whether these glycolytic targets are less dependent on MondoA in HA1ER cells than our previous experiments in C2C12 cells suggest (4) or whether the partial knockdown of MondoA in this experiment was insufficient to reveal a strong MondoA dependence at these targets. It will be of interest to determine whether MondoA cooperates with other cell-type-specific transcriptional regulators to control 2-DG-induced gene expression. Finally, it will be important to determine the nature of MondoA:Mlx regulation of target gene expression as it shuttles between the OMM and the nucleus. Does the heterocomplex occupy targets transiently or is activity restricted to glucose-inducible targets?
Our work establishes MondoA as an important regulator of glucose-induced transcription. MondoA has a paralog called the carbohydrate response element binding protein (ChREBP)/MondoB that also mediates glucose-induced transcription. Despite the gross overall functional similarity and extensive sequence identity shared between MondoA and ChREBP (21), several key differences suggest that they are not functionally identical. For example, none of the phospho-acceptor sites implicated in glucose-regulation of ChREBP are conserved in MondoA (22). MondoA is most highly expressed in skeletal muscle (5), whereas ChREBP is most highly expressed in liver (23). The activity of ChREBP:Mlx complexes is estimated to account for >50% of the glucose transcriptional response in liver with the majority of targets encoding enzymes involved in lipogenesis (24). Our experiments suggest a similar prominent role for MondoA:Mlx complexes in regulating glucose-dependent transcription in skeletal muscle. Therefore, current data suggest that ChREBP:Mlx and MondoA:Mlx are glucose sensors in liver and skeletal muscle, respectively, but each transcription factor complex regulates a set of largely nonoverlapping target genes, resulting in glucose utilization appropriate for the metabolic needs of each tissue.
The final feature of our model of how MondoA:Mlx complexes regulate glucose homeostasis is their negative regulation of glucose uptake. TXNIP levels are elevated in prediabetics and diabetics; however, there is no apparent genetic variation in the TXNIP gene associated with type 2 diabetes (17). Overexpression of TXNIP in skeletal muscle down-regulates peripheral glucose uptake, which is proposed to lead to elevated circulating glucose levels, especially in cases of energy overload (17, 25). Our work suggests that MondoA:Mlx complexes likely account for the previously described transcriptional induction of TXNIP by glucose and the TXNIP-dependent down-regulation of glucose uptake (18). We suggest that elevation of MondoA protein levels or nuclear activity and corresponding down-regulation of glucose uptake may be a contributing factor to the genesis of type 2 diabetes. If so, inhibition of MondoA:Mlx transcriptional activity provides an attractive strategy for the development of antidiabetic therapeutics.
Materials and Methods
Cell Culture.
Cells were maintained at 37°C in 5% CO2 in medium containing penicillin/streptomycin, glutamine (Invitrogen), and 10% bovine calf serum (HyClone) unless otherwise indicated. L6 and HEK293T cells were grown in DMEM, and HA1ER and HA1E cells (gift of William Hahn, Dana-Farber Cancer Institute, Boston) were grown in α-MEM (Mediatech). HA1E cells are human embryonic kidney epithelial cells expressing hTERT and the early region from simian virus 40, whereas HA1ER cells also express the activated H-Ras allele, H-RasG12V (26).
Plasmids and Subcloning.
Plasmids expressing MondoA-V5, Mlx-Flag, and pBabePuroΔN237NLSMondoA have been described (4, 6). MondoA(I766P) and HKII (D657A) were made with the Quikchange XL mutagenesis kit (Stratagene). The TXNIP cDNA was obtained from Open Biosystems.
Sugar and Glucose Analog Treatments.
Cells were incubated overnight in glucose-free DMEM containing 2% serum, followed by treatment in the same medium supplemented with 20 mM of one of the following: glucose, 2-deoxy-d-glucose, 6-deoxy-d-glucose, or 5-thio-d-glucose.
Hexokinase Inhibition and Gene Silencing.
The hexokinase inhibitor, 3BrPA (Aldrich), was used concurrently with 2-DG treatment in glucose-free medium. Nonspecific or MondoA-specific shRNA (M2) were introduced into the HA1ER cells by using the LMP retroviral vector (Open Biosystems) as described (27). The M2 shRNA targets base pairs 2103–2123 of the human MondoA ORF.
Western Blotting.
Primary antibodies were used at the following dilutions: anti-MondoA 1:500 (4), anti-VDUP1 (TXNIP) (Medical and Biological Laboratories) and HKII (Cell Signaling Technology) 1:1,000; antitubulin (Sigma) 1:10,000; and secondary antibodies (Amersham Biosciences and R&D Systems) 1:5,000. Western Lightning Chemiluminescence Plus (Perkin-Elmer) was used for detection.
Immunofluorescence and Microscopy.
Cells were fixed on glass coverslips in 1× PBS containing 3.7% formaldehyde for 10 min and labeled by standard indirect immunofluorescence procedures. Mouse anti-V5 (Invitrogen), rabbit anti-V5 (Sigma), and mouse anti-Flag (Sigma) each were used at 1:1,000. Secondary antibodies (Molecular Probes) were used at 1:500. Hoechst 33342 (2 μg/ml; Molecular Probes) was used to label nuclei. Subcellular localization of immunoreactive MondoA or Mlx protein within individual cells was scored as being intensely cytoplasmic, equally represented between the cytoplasm and nucleus, or more intensely nuclear. Data shown represent the average ± SD resulting from cell counts in five random fields per sample, performed in triplicate.
ChIP.
HA1ER cells were grown in normal or glucose-free medium overnight, then changed into glucose, 2-DG, or glucose-free medium as indicated. ChIP experiments were conducted essentially as described (4) with normalization against a non-E box-containing segment of the BCL-XL promoter.
Expression Analysis.
HA1ER cells at 50% confluence were incubated overnight in regular growth medium or in glucose-free medium, treated for 3 h with either 20 mM glucose or 2-DG, followed by total RNA extraction using RNeasy Mini Kit (Qiagen). cDNA was generated from 2 μg of total RNA by using the SuperScript III RT system (Invitrogen).
Quantitative PCR (qPCR).
qPCR was performed by using iQ SYBR Green Supermix, iCycler, and iCycler software version 3.0a (Bio-Rad) to quantify RNA levels from the expression experiments and DNA levels from the ChIP experiments. Measurements represent the average and SD of two biological replicates for RNA analysis and the average of triplicate biological replicates and SEM for the ChIP experiments. Primer sequences are available on request.
Gene Expression Profiling.
Control or MondoA knockdown HA1ER cells were plated in quadruplicate for glucose starvation, 2-DG treatments, and subsequent isolation of total RNA as described. Total RNA was amplified, Cy3/Cy5-labeled, and hybridized to 44k expression array slides (Agilent) by the Huntsman Cancer Institute Microarray Core Facility. Hybridization was confirmed by using Agilent internal controls, and data were analyzed by using Genesifter (VizX Labs).
Supplementary Material
Acknowledgments.
We thank Sandhya Ravichandran for technical assistance and Steve Lessnick and members of D.E.A.'s laboratory for reviewing the manuscript. This work was supported by Developmental Biology Training Grant T32 HD007491 (to C.W.P.), National Institutes of Health Grants GM55668 and GM60387 (to D.E.A.), and the Huntsman Cancer Foundation. DNA sequencing and oligonucleotide synthesis were supported by Cancer Center Support Grant 2P30 CA42014.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/GEO (accession no. GSE11242).
This article contains supporting information online at www.pnas.org/cgi/content/full/0712199105/DCSupplemental.
References
- 1.Herman MA, Kahn BB. Glucose transport and sensing in the maintenance of glucose homeostasis and metabolic harmony. J Clin Invest. 2006;116:1767–1775. doi: 10.1172/JCI29027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pedersen PL. Warburg, me and Hexokinase 2: Multiple discoveries of key molecular events underlying one of cancers' most common phenotypes, the “Warburg Effect”, i.e., elevated glycolysis in the presence of oxygen. J Bionenerg Biomembr. 2007;39:211–222. doi: 10.1007/s10863-007-9094-x. [DOI] [PubMed] [Google Scholar]
- 3.Freyssenet D. Energy sensing and regulation of gene expression in skeletal muscle. J Appl Physiol. 2007;102:529–540. doi: 10.1152/japplphysiol.01126.2005. [DOI] [PubMed] [Google Scholar]
- 4.Sans CL, Satterwhite DJ, Stoltzman CA, Breen KT, Ayer DE. MondoA-Mlx heterodimers are candidate sensors of cellular energy status: Mitochondrial localization and direct regulation of glycolysis. Mol Cell Biol. 2006;26:4863–4871. doi: 10.1128/MCB.00657-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Billin AN, Eilers AL, Coulter KL, Logan JS, Ayer DE. MondoA, a novel basic helix–loop–helix-leucine zipper transcriptional activator that constitutes a positive branch of a max-like network. Mol Cell Biol. 2000;20:8845–8854. doi: 10.1128/mcb.20.23.8845-8854.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eilers AL, Sundwall E, Lin M, Sullivan AA, Ayer DE. A novel heterodimerization domain, CRM1, and 14–3-3 control subcellular localization of the MondoA-Mlx heterocomplex. Mol Cell Biol. 2002;22:8514–8526. doi: 10.1128/MCB.22.24.8514-8526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li MV, Chang B, Imamura M, Poungvarin N, Chan L. Glucose-dependent transcriptional regulation by an evolutionarily conserved glucose-sensing module. Diabetes. 2006;55:1179–1189. doi: 10.2337/db05-0822. [DOI] [PubMed] [Google Scholar]
- 8.Chi MM, et al. Enzymatic assays for 2-deoxyglucose and 2-deoxyglucose 6-phosphate. Anal Biochem. 1987;161:508–513. doi: 10.1016/0003-2697(87)90481-7. [DOI] [PubMed] [Google Scholar]
- 9.Wilson JE. Isozymes of mammalian hexokinase: Structure, subcellular localization, and metabolic function. J Exp Biol. 2003;206:2049–2057. doi: 10.1242/jeb.00241. [DOI] [PubMed] [Google Scholar]
- 10.Mathupala SP, Ko YH, Pedersen PL. Hexokinase II: Cancer's double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene. 2006;25:4777–4786. doi: 10.1038/sj.onc.1209603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones AR, Gillan L, Milmlow D. The anti-glycolytic activity of 3-bromopyruvate on mature boar spermatozoa in vitro. Contraception. 1995;52:317–320. doi: 10.1016/0010-7824(95)00217-x. [DOI] [PubMed] [Google Scholar]
- 12.Arora KK, Filburn CR, Pedersen PL. Glucose phosphorylation: Site-directed mutations which impair the catalytic function of hexokinase. J Biol Chem. 1991;266:5359–5362. [PubMed] [Google Scholar]
- 13.Shamji AF, Nghiem P, Schreiber SL. Integration of growth factor and nutrient signaling: Implications for cancer biology. Mol Cell. 2003;12:271–280. doi: 10.1016/j.molcel.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 14.Patwari P, Higgins LJ, Chutkow WA, Yoshioka J, Lee RT. The interaction of thioredoxin with Txnip: Evidence for formation of a mixed disulfide by disulfide exchange. J Biol Chem. 2006;281:21884–21891. doi: 10.1074/jbc.M600427200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto Y, et al. Cloning and characterization of a novel gene, DRH1, down-regulated in advanced human hepatocellular carcinoma. Clin Cancer Res. 2001;7:297–303. [PubMed] [Google Scholar]
- 16.Chung JW, Jeon JH, Yoon SR, Choi I. Vitamin D3 up-regulated protein 1 (VDUP1) is a regulator for redox signaling and stress-mediated diseases. J Dermatol. 2006;33:662–669. doi: 10.1111/j.1346-8138.2006.00156.x. [DOI] [PubMed] [Google Scholar]
- 17.Parikh H, et al. TXNIP regulates peripheral glucose metabolism in humans. PLoS Med. 2007;4:e158. doi: 10.1371/journal.pmed.0040158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minn AH, Hafele C, Shalev A. Thioredoxin-interacting protein is stimulated by glucose through a carbohydrate response element and induces β-cell apoptosis. Endocrinology. 2005;146:2397–2405. doi: 10.1210/en.2004-1378. [DOI] [PubMed] [Google Scholar]
- 19.Cho YH, Yoo SD, Sheen J. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell. 2006;127:579–589. doi: 10.1016/j.cell.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 20.Santangelo GM. Glucose signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:253–282. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Billin AN, Ayer DE. The Mlx network: Evidence for a parallel Max-like transcriptional network that regulates energy metabolism. In: Eisenman RN, editor. The Myc/Max/Mad Transcription Factor Network. Heidelberg: Springer; 2006. pp. 255–278. [DOI] [PubMed] [Google Scholar]
- 22.Uyeda K, Repa JJ. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab. 2006;4:107–110. doi: 10.1016/j.cmet.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Postic C, Dentin R, Denechaud PD, Girard J. ChREBP, a transcriptional regulator of glucose and lipid metabolism. Annu Rev Nutr. 2007;27:179–192. doi: 10.1146/annurev.nutr.27.061406.093618. [DOI] [PubMed] [Google Scholar]
- 24.Ma L, Robinson LN, Towle HC. ChREBP*Mlx is the principal mediator of glucose-induced gene expression in the liver. J Biol Chem. 2006;281:28721–28730. doi: 10.1074/jbc.M601576200. [DOI] [PubMed] [Google Scholar]
- 25.Muoio DM. TXNIP links redox circuitry to glucose control. Cell Metab. 2007;5:412–414. doi: 10.1016/j.cmet.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 26.Hahn WC, et al. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol. 2002;22:2111–2123. doi: 10.1128/MCB.22.7.2111-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dickins RA, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.