Abstract
The rhizobia–legume, root-nodule symbiosis provides the most efficient source of biologically fixed ammonia fertilizer for agricultural crops. Its development involves pathways of specificity, infectivity, and effectivity resulting from expressed traits of the bacterium and host plant. A key event of the infection process required for development of this root-nodule symbiosis is a highly localized, complete erosion of the plant cell wall through which the bacterial symbiont penetrates to establish a nitrogen-fixing, intracellular endosymbiotic state within the host. This process of wall degradation must be delicately balanced to avoid lysis and destruction of the host cell. Here, we describe the purification, biochemical characterization, molecular genetic analysis, biological activity, and symbiotic function of a cell-bound bacterial cellulase (CelC2) enzyme from Rhizobium leguminosarum bv. trifolii, the clover-nodulating endosymbiont. The purified enzyme can erode the noncrystalline tip of the white clover host root hair wall, making a localized hole of sufficient size to allow wild-type microsymbiont penetration. This CelC2 enzyme is not active on root hairs of the nonhost legume alfalfa. Microscopy analysis of the symbiotic phenotypes of the ANU843 wild type and CelC2 knockout mutant derivative revealed that this enzyme fulfils an essential role in the primary infection process required for development of the canonical nitrogen-fixing R. leguminosarum bv. trifolii-white clover symbiosis.
Keywords: nitrogen fixation, nodulation, clover, root hair, cellulose
A central event in development of the Rhizobium–legume root-nodule symbiosis is the localized erosion of a cellulosic plant wall through which the bacterial symbiont passes to establish a nitrogen-fixing, intracellular endosymbiotic state within its legume host. Plant cell wall-degrading enzymes are predicted to participate in two steps of this infection process: during primary infection of host root hairs leading to infection thread formation (Inf) and later during bacterial release (Bar) from infection threads within host nodule cells. This process of plant cell wall degradation must be delicately balanced to allow the localized penetration of the bacterial symbiont into the host cell without its overt lysis and destruction.
Several studies indicate that rhizobia produce enzymes capable of degrading plant cell wall polymers (1–14), but little is known about their molecular properties, none have previously been purified to homogeneity, and their specific role (if any) in symbiosis is undefined. The relatively low activities of these rhizobial enzymes have hampered research progress in this area. Using improved assays with increased sensitivity that reliably detect these enzyme activities, we established that cellulases are produced by wild-type strains of Rhizobium leguminosarum (biovars trifolii, phaseoli, and viciae), Bradyrhizobium japonicum, Mesorhizobium loti, and Sinorhizobium meliloti (7, 9). Further studies using R. leguminosarum bv. trifolii ANU843 indicated that this model wild-type strain of clover-nodulating rhizobia produces at least two cellulase isozymes, C1 and C2 (9). Several lines of evidence support the interpretation that both enzymes are cell-bound: (i) their activity is detected in extracts after lysozyme-EDTA treatment or brief sonication of washed and pelleted cells but is not detected in the extracellular culture medium (7, 9), (ii) both scanning and transmission electron microscopy reveal eroded concave pits that replicate the in situ contour of individual bacterial cells attached to root epidermal cell walls of white clover (10), and (iii) data reported here show that the amino acid sequence encoded by the celC2 gene in ANU843 contains a leader signal peptide with several distinctive features used in the general Sec-dependent export pathway of cell envelope proteins. Both C1 and C2 isozymes are made constitutively by ANU843 when grown in defined B-INOS medium, and their approximate molecular masses are 41.5 and 33.2 kDa, respectively (9). Studies using the symbiotic plasmid (pSym)-cured and nod-recombinant derivatives of ANU843 indicated that gene(s) required for production of cellulase C1 are located in pSym but outside its 14-kb HindIII nod region, whereas the locus for the cellulase C2 is not pSym-borne (6).
Recently, a combination of phase-contrast/polarized-light microscopy and enzymology indicated that these cell-bound enzymes from R. leguminosarum bv. trifolii can completely erode through the white clover root hair wall at a highly localized site on the isotropic, noncrystalline apex of its root-hair tip [expression of the Hot (Hole on the tip) phenotype], and can more extensively degrade isolated clover root-hair walls as a substrate if they were grown in the presence (rather than the absence) of purified chitolipooligosaccharide Nod factors from clover rhizobia (10). Other studies showed that these Nod factors from clover rhizobia evoked a localized disruption in the normal crystallization of the host cell wall architecture in growing root hairs (15). Considered collectively, the results implicate a complimentary role of Rhizobium cellulase and Nod factors in promoting root-hair infectibility at strategic sites during primary-host infection. In the present study, we have purified one of the R. leguminosarum bv. trifolii cell-bound cellulase isozymes (CelC2) to homogeneity and analyzed its symbiotic function by a combination of biochemical, reverse genetics, and plant-microscopy approaches. The results provide compelling evidence that this enzyme fulfils a very significant role in the primary-infection process required for development of the canonical nitrogen-fixing, R. leguminosarum bv. trifolii-white clover symbiosis.
Results and Discussion
Cellulases in Rhizobia.
Currently, the known diversity of bacteria capable of forming a nitrogen-fixing root-nodule symbiosis with legumes includes 10 genera and >50 species. All of the official type strains of each of these taxa were examined for cellulase activity, and all were found to be positive to varying degree [supporting information (SI) Table S1]. Evaluation of the currently defined genomes of rhizobia indicated genes coding for putative cellulases represented by a diversity of glycosyl hydrolase families (Table S2). Interestingly, these endoglucanase-coding genes found in rhizobial genomes are located near putative cellulose synthase genes.
Purification and Biochemical Characterization of Cellulase CelC2 from Wild-Type R. leguminosarum bv. trifolii ANU843.
The two cellulase isozymes in the crude extract of ANU843 sonicated cells were resolved by activity gel electrophoresis and were named CelC1 and CelC2 (Fig. S1, lane 6). Anionic exchange chromatography using DEAE Sepharose CL6B separated these two cellulase isozymes, with CelC1 eluting in the peak fraction at 0.1 M NaCl and CelC2 in the peak fraction at 0.2 M NaCl (57.0 units; 0,7 units/mg). Gel-filtration chromatography of the second DEAE peak fraction through Sephacryl S-100 HR resolved one peak of cellulolytic activity (24.0 units; 4.1 units/mg) with an apparent molecular mass of 33.2 kDa, consistent with the results of the activity gel-electrophoresis step indicating that the second DEAE peak contained only one cellulase isozyme (CelC2). Ion-exchange FPLC of this S-100 fraction resolved one peak of cellulolytic activity (6.7 units; 41.9 units/mg) that eluted at 0.2 M NaCl. SDS/PAGE and activity gel electrophoresis of this Mono Q fraction revealed four different proteins, one of which had cellulolytic activity with the same mobility as a silver-stained protein band with an apparent molecular mass of 33.2 kDa (Fig. S1, lane 4). Dialysis of this Mono Q fraction, followed by hydrophobic-interaction FPLC using a Phenyl Superose column separated four protein peaks, one of which eluted at 0.26 M ammonium sulfate and contained all of the cellulolytic activity (5.0 units; 152 units/mg) of the applied sample (Fig. S1, lane 7).
Final purification of the CelC2 cellulase isozyme to homogeneity was indicated by a single peak of protein with constant cellulase-specific activity using gel filtration through Sephacryl S-200 and a single band of silver-staining protein in SDS/PAGE that matched the electrophoretic mobility of cellulase CelC2 detected by activity gel electrophoresis of the final purified enzyme (Fig. S1, lanes 5 and 7). These results indicate that active, purified CelC2 cellulase from R. leguminosarum bv. trifolii ANU843 consists of a single peptide with molecular mass of 33.2 kDa. Its hydrolytic enzyme activity using carboxymethyl cellulose (CMC) as substrate has an optimal pH of 5.0, an optimal temperature of 40°C, a specific activity of 152 units/mg of protein and an apparent Km of 84.4 mg/ml.
Purified CelC2 cellulase hydrolyzed CMC with the release of reducing sugar equivalents; but, under the same conditions, this purified enzyme did not release reducing sugars from Avicel microcrystalline cellulose, cellobiose, polygalacturonate, locust bean gum, xylan, R. leguminosarum bv. trifolii acidic heteropolysaccharide EPS types I, II, or III, or S. meliloti succinoglycan exopolysaccharide. These data indicate that purified CelC2 cellulase is a 1,4-β-d-endoglucanase with high substrate specificity for noncrystalline cellulose, and, unlike R. leguminosarum PlyA and PlyB glycanases (3, 16), CelC2 does not degrade any of the three different chemotypes of extracellular acidic heteropolysaccharides made by R. leguminosarum bv. trifolii. Also, the substrate specificity of ANU843 CelC2 does not extend to the β-1,4 heptaglucose backbone in the repeated octasaccharide of S. meliloti succinoglycan.
Molecular Analysis of CelC2.
An Edman degradation of purified ANU843 CelC2 cellulase revealed that its N terminus is blocked. An internal peptide isolated by RP-HPLC from a tryptic digestion of purified CelC2 contained 14 amino acids with a sequence of Ala-Glu-Gly-Phe-Asp-Ala-Glu-Phe-Gly-Tyr-Asn-Ala-Ile-Arg, which has 85% identity to a sequence encoded by the celC gene of R. leguminosarum bv. trifolii R200 (17). The celC-encoded protein has sequence homology to endoglucanase CelC from Agrobacterium tumefaciens (18), CMCax from Gluconacetobacter xylinus (19), CelY from Erwinia chrysanthemi (20), Cuda from Cellulomonas uda (21), and BcsZ from Escherichia coli (22), all belonging to the glycosyl hydrolase family 8 (23).
Identification and Cloning of the CelC2-Encoding Gene.
The primers CelCexF and C2R were used for PCR amplification of celC2 using DNA from strain ANU843 as template. The amplified DNA fragment contained a 1,044-bp ORF (GenBank accession no. AJ561043) encoding a predicted protein of 347 aa with a Sec-dependent (General Secretory Pathway) signal peptide of 23 aa (Fig. S2). The amino acid sequence of the putative celC2-encoded protein contains the sequence of the tryptic peptide isolated from the purified enzyme, confirming that the amplified sequence matched that of our target gene (Fig. S2). This celC2 gene from R. leguminosarum bv. trifolii ANU843 has 88% identity to cel8A from R. leguminosarum bv. trifolii 1536 (GenBank accession no. AY227046) (24), 86% to celC from R. leguminosarum bv. trifolii R200 (GenBank accession no. AF121340) (17), 86% to celY from R. leguminosarum bv. viciae 3841 (GenBank accession no. AM236080) (25), 85% to celC from R. etli CFN42 (GenBank accession no. CP000133) (26), and 69% to Smed_5210 from Sinorhizobium medicae WSM419 (GenBank accession no. CP000740).
Linkage of Genes Encoding Cellulase and Cellulose Biosynthesis Functions in Rhizobia.
Orthologs to the celC endoglucanase-coding gene are found in a diversity of eubacteria that make cellulose (Rhizobium, Agrobacterium, Gluconacetobacter, E. coli, Salmonella, and others). In all of these cellulose-producing eubacteria, the celC genes are located near putative cellulose synthase genes localized in a region of the chromosome (celABC or bcsABZ) involved in bacterial cellulose biosynthesis (17–19). In the currently defined rhizobial genomes where the celC gene is present (R. leguminosarum, R. etli, and S. medicae), this gene is located in this cellulose synthase operon. The celC2 from R. leguminosarum bv. trifolii ANU843 is also located in this cellulose synthase operon (Fig. S2). The primary sequence and organization of the celABC genes are similar in R. leguminosarum bv. trifolii R200 (17) and A. tumefaciens (18). The postulated involvement of the Agrobacterium CelC cellulase enzyme in bacterial cellulose biosynthesis is to transfer new oligomer primers for chain elongation (27), but that biochemical function has not been definitively established. The lack of cellulose biosynthesis by mutation of celA or celB of R. leguminosarum bv. trifolii did not affect its ability to nodulate clover under controlled laboratory conditions (17). Nevertheless, it is feasible that the benefit to rhizobia gain by their cellulose microfibril-facilitated firm adhesion during colonization of host roots (28, 29) has significant symbiotic importance under natural conditions in rhizosphere soil.
Biological Activity of Purified Cellulase CelC2 from R. leguminosarum bv. trifolii ANU843 on White Clover Seedling Roots.
Previous studies (10) showed that the DEAE fraction containing the partially purified cellulase CelC2 isozyme from wild-type ANU843 was capable of degrading the cell wall at the apex of the root hair tip (the “Hot” phenotype) in vivo when incubated with intact white clover host-seedling roots, and this activity was not found in the other DEAE fraction containing the cellulase C1 isozyme (10). The symbiotic relevance of this enzyme bioactivity was revealed by studies indicating that (i) its highly localized site of cell wall degradation matched the restricted distribution of the isotropic noncrystalline wall architecture at root hair tips of axenic white clover seedlings that typically undergo marked curling and infection (10), (ii) the discrete hole through the plant wall that is ultimately produced there has a diameter precisely within the 2- to 3-μm range typically found at primary infection sites in deformed root hairs (10, 30, 31), (iii) it was more extensive in walls of root hairs that had been grown in the presence of wild-type chitolipooligosaccharide Nod factors that increase root hair infectibility (10), and (iv) it was less extensive when clover seedlings were grown with sufficient nitrate supply to inhibit primary infection (10, 32) or with root hair walls isolated from the nonhost legume, alfalfa (15).
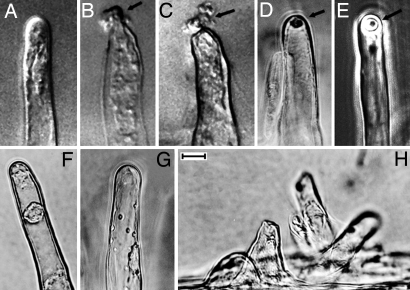
To test whether CelC2 cellulase exhibits this bioactivity, we performed this direct in situ assay using the fully purified isozyme from wild-type ANU843 on axenic seedling roots of its white clover host. As predicted, the results indicated that the purified CelC2 cellulase exhibits this same infection-related bioactivity as the previously described, partially purified DEAE fraction containing it. The series of images in Fig. 1 show a progression of the morphological features of the Hot phenotype produced in vivo on susceptible root hairs treated with purified CelC2 (Fig. 1 A–E). The enzyme acting at this localized site erodes a hole completely through the root-hair wall, resulting in the extrusion of its protoplast/cytoplasm (Fig. 1 B and C). The Hot phenotype occurred at an approximate frequency of 14 root hairs per centimeter of optical median plane on axenic white clover seedlings after 6 h of enzyme incubation. No Hot activity was found on root hairs of clover seedlings incubated with the isotonic Fähraeus medium without enzyme as a negative control (Fig. 1A) or when the purified CelC2 enzyme from ANU843 was incubated with axenic seedling roots of the nonhost legume, alfalfa (Fig. 1G). As a positive control, incubation of both white clover and alfalfa seedlings with a commercial fungal cellulase from Trichoderma (Sigma) under the same experimental conditions resulted in extensive degradation of root hairs (Fig. 1H and data not shown). These results provide direct, in situ evidence clearly indicating that purified cellulase CelC2 from ANU843 can degrade the clover host-plant cell wall precisely at the strategic location where the bacterial symbiont first penetrates (markedly curled) root hairs and suggests that the specificity of that enzyme-catalyzed reaction may contribute to the restricted host range of root-hair infection expressed in the Rhizobium–clover symbiosis.
Fig. 1.
Biological activity of purified cellulase CelC2 from R. leguminosarum bv trifolii ANU843 on clover root hairs. (A–C) Nomarski interference contrast micrographs. (A) Typical control root hair without enzyme treatment. (B) Early stage in expression of the Hot phenotype. A portion of the root hair protoplast (arrow) has extruded through the hole made at the tip of the root hair wall by CelC2. (C) Further extrusion of a larger portion of the protoplast (arrow) through the hole made by CelC2 cellulase at a root-hair tip. (D–H) Phase-contrast micrographs. (D and E) The refractile hole (arrow) made by CelC2 cellulase at the tip of a clover root hair after its cytoplasm has already been expelled into the external rooting medium. (F and G) Alfalfa root hairs remain intact in the untreated control (F) and after incubation with CelC2 from ANU843 (G). (H) Extensive degradation of clover root hairs after treatment with a commercial fungal cellulase. (Scale bar: 12 μm.)
Symbiotic Phenotype of an ANU843 celC2 Knockout Mutant Derivative Strain.
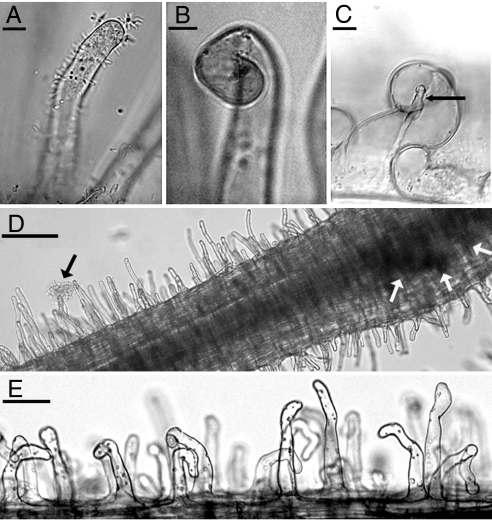
To further examine whether the activities of CelC2 are infection-related, we compared the symbiotic phenotypes of wild-type ANU843 to the C2-cellulase knockout mutant derivative ANU843ΔcelC2 obtained by an internal 361-bp BamHI-HindIII fragment deletion that encodes the GH8 glycosyl hydrolase catalytic center motif within the celC2 gene (Fig. S2). The wild-type strain accomplished the canonical steps in the infection process with white clover: root-hair adhesion (Fig. 2A), various types of root-hair deformation (Had, Fig. 2B), marked root hair curling (Hac, “shepherd crook”) and infection-thread formation (Inf, Fig. 2C). The ANU843 ΔcelC2 mutant retained the ability to adhere to the host root surface (Fig. 2D) and induced various types of moderate root-hair deformation (Fig. 2E) but did not evoke formation of complete, marked root hair shepherd crooks or complete infection threads when inoculated onto white clover seedlings (Fig. 2 and Table S3).
Fig. 2.
Primary-infection events in white clover inoculated with wild-type ANU843 (A–C) and the CelC2− mutant ΔcelC2 (D and E). The phase-contrast micrographs show the canonical steps in primary host infection of white clover by R. leguminosarum bv trifolii; root hair adhesion (A), markedly curled “shepherd crook” (B), and infection-thread formation (C, arrow). The CelC2− mutant ΔcelC2 exhibits root-hair adhesion (D, black arrow) and induces root hair deformation (E) and nodule initiation (D, white arrows) but no infection threads in white-clover root hairs (D and E). (Scale bars: 15 μm, A–C; 50 μm, E; and 100 μm, D.)
Detailed examination by phase-contrast microscopy of the seedlings inoculated with the ANU843ΔcelC2 mutant indicated only two rare cases where a localized refractile bright spot developed on deformed root hairs. That morphological feature commonly precedes root-hair penetration and infection-thread formation (15). As is also the case for root hairs responsive to Nod factors (15), a subset of root hairs is particularly susceptible to deformation after inoculation with either the wild type or the CelC2− mutant derivative strain (Fig. 2E). The most susceptible root hairs were actively growing in the root hair zones I and II. Root hairs that have finished growing (root hair zone III) were refractory to deformation.
Both ANU843 and ΔcelC2 test strains induced nodule formation on white clover roots (Fig. 2D and Table S3). The first nodules emerged 10 days after inoculation with either strain, and all inoculated plants were nodulated by the third week. The mean number of nodules per plant at 40 days after inoculation was higher with the mutant than with the wild-type strain, but the difference in mean values was not statistically significant (Table S3). Increased nodulation is a common feature for ineffective Rhizobium–legume symbioses (33).
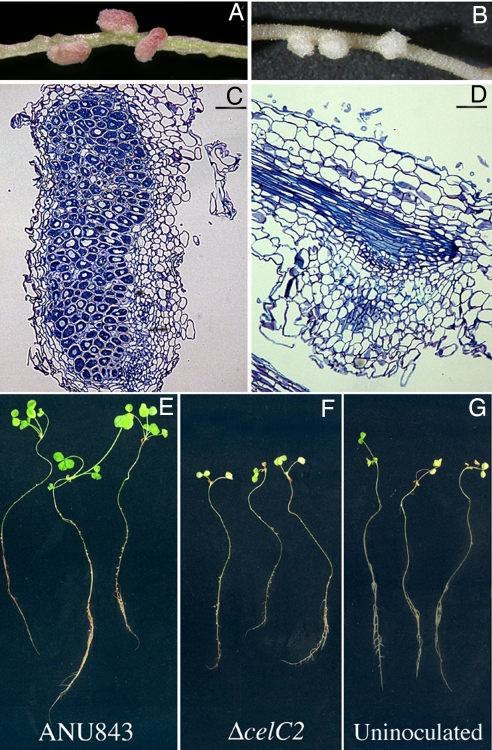
The nodule organogenesis in the inner cortex (Noi phenotype) was similar in white clover plants inoculated with the wild-type ANU843 or the CelC2 minus strain (Table S3 and Fig. 2D). Emerged nodules induced by the wild type were invaded, typically red, piriform, and indeterminate (Fig. 3 A and C). In contrast, emerged nodules induced by the mutant ΔcelC2 were small, white, spherical, and uninvaded (Fig. 3 B and D).
Fig. 3.
Nodule development (Nod) in white clover 40 days after inoculation with the wild-type ANU843 (A, C, and E) or the CelC2− mutant ΔcelC2 (B, D, and F); shown are portions of nodulated roots from inoculated plants (A and B), longitudinal sections of the induced nodules (C and D), and whole-plant phenotypes (E–G). (Scale bars: 200 μm.)
Plants grew well in the nitrogen-free medium when inoculated with the wild-type strain ANU843, indicative of an effective nitrogen-fixing symbiosis. Those plants benefitted by the symbiosis produced greener leaves and shoots that were significantly longer than plants inoculated with the mutant ΔcelC2 or the uninoculated axenic control plants (Table S3 and Fig. 3 E–G). The latter two groups of plants were stunted in development and became chlorotic once the fixed nitrogen from their cotyledons was exhausted, indicative of nitrogen-starvation stress without nitrogen fixation under these N-free growth conditions. Complementation of the mutant strain ANU843ΔcelC2 with a plasmid expressing the celC2 gene restored the ability to produce the CelC2 cellulase isoenzyme, infect root hairs, and induce efficient nitrogen-fixing nodules on white clover (Fig. S3).
To the best of our knowledge, the biochemical characteristics and infection-related biological activity of a purified rhizobial enzyme capable of creating the hollow, localized portal of entry into the root hair cell wall through which the bacterial symbiont can penetrate during primary infection of its legume host has not been previously reported. Our purification protocol yields a cell-bound β-1,4-endoglucanase (EC 3.2.1.4) isozyme from the clover microsymbiont, R. leguminosarum bv. trifolii. This enzyme, called CelC2 cellulase, is particularly important for symbiotic development because this rhizobial symbiont requires its activity to breach the host barrier to establish an endosymbiotic, nitrogen-fixing association with its specific legume host.
Further studies reported here and elsewhere revealed multiple characteristics of CelC2 cellulase that predictively restrict (thereby tightly control) its symbiotically-relevant activity during primary-host infection. First, the cell-bound location (7, 9), relatively low quantity produced (rather than excretion of large quantities into the external environment), and high Km relative to other microbial cellulases are all characteristics that would predictively restrain its degradative action on roots, thereby minimizing indiscriminate and overextended wall hydrolysis inevitably resulting in host-cell lysis and death. Elevated levels of root hair-wall peroxidase localized at this same site likely play an important complementary role in maintaining the viability of the host cell during incipient microsymbiont penetration (34). Second, these same characteristics would provide further opportunity to restrict its short-range action based on physical positioning of the bacterium at the host-wall interface. Third, the substrate specificity of CelC2 cellulase for noncrystalline cellulose significantly restricts its in vivo site of wall-penetrating erosive action to the highly localized root-hair infection sites lacking crystalline wall architecture. This finding provides an explanation of the remarkable infection event that rhizobia typically only erode and penetrate one side of the root-hair wall despite being sandwiched by both sides when cradled in the overlap of the markedly curled shepherd's crook (10, 15, 29, 30, 35, 36). Development of these localized infection sites is strategically modulated by the action of the rhizobial chitolipooligosaccharide Nod factors that disrupt the crystallization of the root-hair wall architecture and activate symbiotic root-hair infection (15). Fourth, the diameter of the hole eroded completely through the root-hair wall by CelC2 cellulase action is precisely within the same in situ size range that wild type R. leguminosarum bv trifolii cells use to traverse the clover root-hair wall during primary host infection (10, 30, 31). Fifth, the pH 5 optimum for CelC2 cellulase is compatible with the slightly acidic pH measured in vivo at the external surface of white clover root hairs (J. Salzwedel and F.D., unpublished data). All of these biochemical and cytological properties of CelC2 cellulase are typical of what one would expect for a Rhizobium plant wall-degrading enzyme that plays an important contributing role in creating the unique portal of microsymbiont entry during early steps of primary-host infection required for development of the endosymbiotic nitrogen-fixing Rhizobium–white clover symbiosis. Finally, CelC2 knockout mutants were unable to breach the host wall at the root-hair tip and form infection threads that are necessary to invade nodules allowing them to fix nitrogen, and transfer of the cloned wild-type gene into the CelC2 knockout mutant restored these symbiotic phenotypes, providing further compelling evidence for the requirement of this enzyme in successful development of the canonical Rhizobium–white clover symbiosis.
Materials and Methods
Bacterial Strains and Growth Conditions.
Wild-type R. leguminosarum bv. trifolii strain ANU843 (from B. Rolfe, Australian National University, Canberra, Australia) and its derivative ANU843ΔcelC2 were grown in defined B-INOS (9) or yeast mannitol agar (YMA) media (37) at 28°C. These media were supplemented with kanamycin (50 μg/ml) for growth of the celC2-complemented strain.
DNA Methods and Construction of ANU843 Derivative Strains.
DNA was extracted according to Rivas et al. (38). Plasmid DNA was obtained by using a High Pure Plasmid Isolation kit (Roche Molecular Biochemicals) by following manufacturer's instructions. Preparation of plasmid DNA, restriction enzyme digestions, DNA cloning, PCR DNA amplification, and agarose DNA electrophoresis were performed by using standard procedures (39).
To generate the ΔcelC2 deletion mutant derivative, CelCexF (TCGCCGCCAACTGGCTGTC) and C2R (CACAGACACTCCGGATGC) primers were used to amplify celC2 from ANU843 DNA. The PCR product was cloned into pGEM-T Easy vector (Promega) and a 361-bp BamHI-HindIII fragment from celC2 was removed. The truncated celC2 gene was cloned as a PvuII fragment into plasmid pK18mobsacB (40) and transferred to R. leguminosarum bv. trifolii ANU843 by triparental mating using pRK2013 as helper plasmid (41). Allele replacement was selected as described (40). PCR analysis with primers C1F (ATCAGCCACAGCGAAGGGCA) and C2R was used to verify the 361-bp deletion in the mutant strain (Fig. S3A). The lack of CelC2 activity in this deletion mutant was confirmed by a zymogram (Fig. S3B).
To generate the complemented strain, an ANU843 genomic region containing the CelC2 coding sequence and 78 nt upstream of the start codon was amplified as a SalI fragment and inserted between the XhoI and SalI sites into plasmid pBBRMS-2 (42) to yield pJZC2, which was transferred to R. leguminosarum bv. trifolii ANU843ΔcelC2 by conjugation as described.
Enzyme Assays.
CMCase activities were detected in vitro by the double-layer plate enzyme assay using Congo red staining (9), reducing sugar assays using 2,2′-bicinchonic acid (BCA) (43), and the activity stain overlay technique using SDS/PAGE, followed by renaturation (9). One unit of enzyme activity is defined as the amount releasing 1 nmol of reducing sugar equivalent per min at 40°C. One unit of specific activity was defined as 1 nmol reducing sugar equivalent released min−1 (mg protein)−1. The amount of protein was measured by the dye-binding method of Bradford (44) with BSA as the standard.
Enzyme Purification.
All purification steps were performed at 4°C, and protein profiles in chromatographic fractions were analyzed by SDS/PAGE, followed by silver staining (45). Broth cultures (12 liters) of wild-type ANU843 in the early stationary phase (9 × 108 cells per ml) were centrifuged at 20,000 × g for 15 min. Pelleted cells were washed by centrifugation with water, suspended in 30 ml of 10 mM Tris·HCl buffer (pH 8.0) on ice, sonicated in five cycles of 1-min bursts at 32 W with a microprobe, and centrifuged at 12,000 × g for 20 min. This sonicated “crude extract” was applied to a DEAE-Sepharose CL-6B column (18 × 3 cm) previously equilibrated with 10 mM Tris·HCl buffer. Unbound protein was washed through the column with 10 mM Tris·HCl buffer. Bound protein was eluted in a linear gradient of 0–0.4 M NaCl in 10 mM Tris·HCl buffer at a flow rate of 1.75 ml/min. Fractions (2.5 ml) containing cellulase activity were pooled and concentrated by ultrafiltration using an Amicon YM 30 membrane, which does not bind these cellulases. This ultrafiltration-concentrated DEAE fraction was chromatographed through a Sephacryl S-100 HR column (95 × 2 cm) by using 100 mM Tris·HCl buffer (pH 8.0, 0.25 ml/min) to avoid nonspecific binding of rhizobial cellulases. The S-100 fractions (2.5 ml) containing cellulase activity were pooled and applied to a Amersham Pharmacia Mono Q HR 5/5 column. Unbound protein was eluted with 10 mM Tris·HCl buffer (pH 8). The bound cellulase activity was eluted by a linear gradient of 0–0.4 M NaCl in 10 mM Tris·HCl buffer at 0.25 ml/min. Mono Q-fractions containing cellulase activity were pooled and mixed with an equal volume of 3.4 M ammonium sulfate in PCA buffer (pH 5.0) to yield a final concentration of 1.7 M ammonium sulfate. This mixture was applied to a Amersham Pharmacia Phenyl-Superose HR 5/5 column equilibrated with 1.7 M ammonium sulfate in 10 mM PCA buffer (pH 5.0, 0.5 ml/min, 0.5-ml fractions). Bound proteins (A280nm) were eluted with a linear gradient of 1.7–0 M ammonium sulfate in 10 mM PCA buffer. Collected fractions (0.5 ml) containing cellulase activity were concentrated by using Centricon YM-10 ultrafilters and stored at 4°C.
Biochemical Characterization of Purified CelC2 Cellulase.
The 1,4-β-d-endoglucanase activity assays were run under standard conditions (9) to determine the optimum pH over the range of 4.0–7.0 (PCA 200 mM) and 8.0–9.0 (Tris·HCl 200 mM) and the influence of temperature on enzyme activity in the range of 4°C to 70°C. The molecular mass of the purified native enzyme was analyzed by gel-filtration chromatography using a Sephacryl S-200 column (100 × 2.5-cm, 2.5-ml fractions) in 100 mM Tris·HCl buffer. The protein content of collected fractions were estimated by A280nm and assayed for enzyme activity (9). The molecular mass was also estimated by activity gel electrophoresis (9). To examine substrate specificity, potential substrates of Avicel crystalline cellulose, cellobiose, Na polygalacturonate, locust bean gum, xylan, R. leguminosarum bv. trifolii EPS types I, II, and III (46), and S. meliloti succinoglycan (each 1 mg/ml in PCA buffer) were compared with CMC under standard assay conditions (9). When necessary, the remaining insoluble substrates were removed by centrifugation (10,000 × g, 15 min) at 4°C before analyzing the supernatant for depolymerized products. To determine the Km for cellulase CelC2, its rate of CMC hydrolysis was measured at substrate concentrations ranging from 0.1% to 1% (wt/vol) at pH 5 and 40°C, followed by graphing its kinetics in a plot of V vs. S and computing its S at 0.5 Vmax. For sequencing analysis, tryptic peptides of the purified enzyme were isolated by using a RP-HPLC column. The automated Applied Biosystems model 477A pulse-liquid sequenator equipment used to perform the Edman degradation was connected on-line to an RP-HPLC unit for identification of the stepwise released PTH-amino acids.
Biological Activity Assay.
Purified CelC2 cellulase (40 μl, 0.2 units) from strain ANU843 was assayed in vivo for its ability to evoke the Hot phenotype (10) on root hairs of intact axenic seedlings of white clover (Trifolium repens L. var. Dutch) and alfalfa (Medicago sativa var. Aragón). Control treatments included nitrogen-free Fähraeus medium (44) without enzyme and commercial fungal cellulase (0.2 units) from Trichoderma (Sigma) in the same medium. Periodically, root hairs were examined individually for localized cell-wall degradation by phase-contrast and Nomarski interference light microscopy at ×400 magnification, using a 546-nm narrow band-pass interference contrast filter to enhance optical refractility by minimizing chromatic aberration.
Analysis of Symbiotic Phenotypes.
Surface-sterilized white clover seeds were germinated for 2 days in humid air on inverted plates of water agar incubated at room temperature in the dark. Five seedlings with straight roots ≈10 mm in length were transferred to the surface of 2% agar plates containing nitrogen-free Fähraeus medium (35) and overlaid with a 9-cm-diameter disk of Whatman No. 1 sterile filter paper. One milliliter of a suspension (OD600 of 0.5) of 5-day-old Rhizobium culture that had been grown on YMA was deposited on the overlying filter paper. Inoculated seedlings were incubated at room temperature in the dark for 5 days and then examined by light microscopy to assess root-hair deformation and infection.
For nodulation phenotypes, axenic seedlings were transferred to a Whatman No. 1 filter paper support (15 × 1.5 cm) placed inside test tubes (one plant per tube) containing 20 ml of nitrogen-free Fähraeus medium. Plants were grown at room temperature in the dark for 5 days and then inoculated (20 per test strain) with 1 ml of a suspension (OD600 of 0.5) of a 5-day-old Rhizobium culture grown on YMA, and transferred to a growth chamber with mixed incandescent and fluorescent illumination (400 microeinsteins m−2 s−1; 400–700 nm) programmed for a 16-h photoperiod, 24°C: 20°C day–night cycle, and 50–60% relative humidity. Roots were examined by stereomicroscopy every 48 h over 40 consecutive days. Then roots and nodules were excised and processed for bright-field light microscopy (47).
Supplementary Material
Acknowledgments.
This work was supported by Ministerio de Educación y Ciencia Grant AGL 2005-07796 from Spain and by the Michigan Agricultural Experiment Station. M.R. was supported by a fellowship from the Ministerio de Educación y Ciencia, Spain. We dedicate this paper in memory of Professor Antonio Palomares, former president of the Spanish Society for Nitrogen Fixation (Sociedad Española de Fijación de Nitrógeno, SEFIN).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ561043).
This article contains supporting information online at www.pnas.org/cgi/content/full/0802547105/DCSupplemental.
References
- 1.Angle JS. Pectic and proteolytic enzymes produced by fast- and slow-growing soybean rhizobia. Soil Biol Biochem. 1986;18:115–116. [Google Scholar]
- 2.Chalifour FP, Benhamou N. Indirect evidence for cellulase production by Rhizobium in pea root nodules during bacteroid differentiation: Cytochemical aspects of cellulose breakdown in rhizobial droplets. Can J Microbiol. 1989;35:821–829. [Google Scholar]
- 3.Finnie C, Zorreguieta A, Hartley NM, Downie JA. Characterization of Rhizobium leguminosarum exopolysaccharide glycanases that are secreted via a type I exporter and have a novel heptapeptide repeat motif. J Bacteriol. 1998;180:1691–1699. doi: 10.1128/jb.180.7.1691-1699.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hubbell DH, Morales VM, Umali-Garcia M. Pectolytic enzymes in Rhizobium. Appl Environ Microbiol. 1978;35:210–213. doi: 10.1128/aem.35.1.210-213.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iannetta P, McMillan G, Sprent JI. Plant cell wall-degrading enzymes of Rhizobium leguminosarum bv. viciae: Their role in avoiding the host-plant defense response. Soil Biol Biochem. 1997;29:1019–1021. [Google Scholar]
- 6.Jiménez-Zurdo JI, Mateos PF, Dazzo FB, Martínez-Molina E. Influence of the symbiotic plasmid (pSym) on cellulase production by Rhizobium leguminosarum bv. trifolii ANU843. Soil Biol Biochem. 1996;28:131–133. [Google Scholar]
- 7.Jiménez-Zurdo JI, Mateos PF, Dazzo FB, Martínez-Molina E. Cell-bound cellulase and polygalacturonase production by Rhizobium and Bradyrhizobium species. Soil Biol Biochem. 1996;28:917–921. [Google Scholar]
- 8.Martínez-Molina E, Morales VM, Hubbell DH. Hydrolytic enzyme production by Rhizobium. Appl Environ Microbiol. 1979;38:1186–1188. doi: 10.1128/aem.38.6.1186-1188.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mateos PF, et al. Cell-associated pectinolytic and cellulolytic enzymes in Rhizobium leguminosarum biovar trifolii. Appl Environ Microbiol. 1992;58:1816–1822. doi: 10.1128/aem.58.6.1816-1822.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mateos PF, et al. Erosion of root epidermal cell walls by Rhizobium polysaccharide-degrading enzymes as related to primary host infection in the Rhizobium–legume symbiosis. Can J Microbiol. 2001;47:475–487. doi: 10.1139/w01-039. [DOI] [PubMed] [Google Scholar]
- 11.Michaud P, Belaich A, Courtois B, Courtois J. Cloning, sequencing and overexpression of a Sinorhizobium meliloti M5N1CS carboxymethyl-cellulase gene. Appl Microbiol Biotechnol. 2002;58:767–771. doi: 10.1007/s00253-002-0953-4. [DOI] [PubMed] [Google Scholar]
- 12.Morales V, Martínez-Molina E, Hubbell D. Cellulase production by Rhizobium. Plant Soil. 1984;80:407–415. [Google Scholar]
- 13.Plazinski J, Rolfe BG. Analysis of the pectolytic activity of Rhizobium and Azospirillum strains isolated from Trifolium repens. J Plant Physiol. 1985;120:181–187. [Google Scholar]
- 14.Verma DPS, Zogbi V, Bal AK. A cooperative action of plant and Rhizobium to dissolve the host cell wall during development of root nodule symbiosis. Plant Science Lett. 1978;13:137–142. [Google Scholar]
- 15.Dazzo FB, et al. Modulation of development, growth dynamics, wall crystallinity, and infection sites in white clover root hairs by membrane chitolipooligosaccharides from Rhizobium leguminosarum biovar trifolii. J Bacteriol. 1996;178:3621–3627. doi: 10.1128/jb.178.12.3621-3627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zorreguieta A, Finnie C, Downie JA. Extracellular glycanases of Rhizobium leguminosarum are activated on the cell surface by an exopolysaccharide-related component. J Bacteriol. 2000;182:1304–1312. doi: 10.1128/jb.182.5.1304-1312.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ausmees N, Jonsson H, Hoglund S, Ljunggren H, Lindberg M. Structural and putative regulatory genes involved in cellulose synthesis in Rhizobium leguminosarum bv. trifolii. Microbiology. 1999;145:1253–1262. doi: 10.1099/13500872-145-5-1253. [DOI] [PubMed] [Google Scholar]
- 18.Matthysse AG, White S, Lightfoot R. Genes required for cellulose synthesis in Agrobacterium tumefaciens. J Bacteriol. 1995;177:1069–1075. doi: 10.1128/jb.177.4.1069-1075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong HC, et al. Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc Natl Acad Sci USA. 1990;87:8130–8134. doi: 10.1073/pnas.87.20.8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guiseppi A, Aymeric JL, Cami B, Barras F, Creuzet N. Sequence analysis of the cellulase-encoding celY gene of Erwinia chrysanthemi: A possible case of interspecies gene transfer. Gene. 1991;106:109–114. doi: 10.1016/0378-1119(91)90573-t. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura K, Misawa N, Kitamura K. Sequence of a cellulase gene of Cellulomonas uda CB4. J Biotechnol. 1986;4:247–254. [Google Scholar]
- 22.Park YW, Yun HD. Cloning of the Escherichia coli endo-1,4-d-glucanase gene and identification of its product. Mol Gen Genet. 1999;261:236–241. doi: 10.1007/s004380050962. [DOI] [PubMed] [Google Scholar]
- 23.Coutinho PM, Henrissat B. The modular structure of cellulases and other carbohydrate-active enzymes: An integrated database approach. In: Ohmiya K, et al., editors. Genetics, Biochemistry and Ecology of Cellulose Degradation. Tokyo: Uni; 1999. pp. 15–23. [Google Scholar]
- 24.An JM, et al. Cloning and characterization of ce/8A gene from Rhizobium leguminosarum bv. trifolii 1536. Lett Appl Microbiol. 2004;38:296–300. doi: 10.1111/j.1472-765x.2004.01485.x. [DOI] [PubMed] [Google Scholar]
- 25.Young JP, et al. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 2006;7:R34. doi: 10.1186/gb-2006-7-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.González V, et al. The partitioned Rhizobium etli genome: Genetic and metabolic redundancy in seven interacting replicons. Proc Natl Acad Sci USA. 2006;103:3834–3839. doi: 10.1073/pnas.0508502103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matthysse AG, Thomas DL, White AR. Mechanism of cellulose synthesis in Agrobacterium tumefaciens. J Bacteriol. 1995;177:1076–1081. doi: 10.1128/jb.177.4.1076-1081.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dazzo FB, et al. Specific phases of root hair attachment in the Rhizobium trifolii–clover symbiosis. Appl Environ Microbiol. 1984;48:1140–1150. doi: 10.1128/aem.48.6.1140-1150.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mateos PF, et al. Direct in situ identification of cellulose microfibrils associated with Rhizobium leguminosarum biovar. trifolii attached to the root epidermis of white clover. Can J Microbiol. 1995;41:202–207. [Google Scholar]
- 30.Callaham DA, Torrey JG. The structural basis for infection of root hairs of Trifolium repens by Rhizobium. Can J Bot. 1981;59:1647–1664. [Google Scholar]
- 31.Napoli C, Dazzo F, Hubbell D. Production of cellulose microfibrils by Rhizobium. Appl Microbiol. 1975;30:123–131. doi: 10.1128/am.30.1.123-131.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dazzo FB, Brill WJ. Regulation by fixed nitrogen of host-symbiont recognition in the Rhizobium–clover symbiosis. Plant Physiol. 1978;62:18–21. doi: 10.1104/pp.62.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fred EB, Baldwin IL, McCoy E. Root nodule bacteria and leguminous plants. Madison, WI: Univ of Wisconsin Press; 1932. [Google Scholar]
- 34.Salzwedel JL, Dazzo FB. pSym nod gene influence on elicitation of peroxidase activity from white clover and pea roots by rhizobia and their cell-free supernatants. Mol Plant–Microbe Interact. 1993;6:127–134. doi: 10.1094/mpmi-6-127. [DOI] [PubMed] [Google Scholar]
- 35.Fähraeus G. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J Gen Microbiol. 1957;16:374–381. doi: 10.1099/00221287-16-2-374. [DOI] [PubMed] [Google Scholar]
- 36.Turgeon BG, Bauer WD. Ultrastructure of infection-threads development during the infection of soybean by Rhizobium japonicum. Planta. 1985;163:328–349. doi: 10.1007/BF00395142. [DOI] [PubMed] [Google Scholar]
- 37.Somasegaran P, Hoben HJ. Handbook for Rhizobia: Methods in Legume-Rhizobium Technology. New York: Springer; 1994. [Google Scholar]
- 38.Rivas R, Velázquez E, Valverde A, Mateos PF, Martínez-Molina E. A two primers random amplified polymorphic DNA procedure to obtain polymerase chain reaction fingerprints of bacterial species. Electrophoresis. 2001;22:1086–1089. doi: 10.1002/1522-2683()22:6<1086::AID-ELPS1086>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 40.Schafer A, et al. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 41.Ditta G, Stanfield S, Corbin D, Helinski DR. Broad host range DNA cloning system for Gram-negative bacteria: Construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kovach ME, Phillips RW, Elzer PH, Roop RM, II, Peterson KM. pBBR1MCS: A broad-host-range cloning vector. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 43.Waffenschmidt S, Jaenicke L. Assay of reducing sugars in the nanomole range with 2,2′-bicinchoninate. Anal Biochem. 1987;165:337–340. doi: 10.1016/0003-2697(87)90278-8. [DOI] [PubMed] [Google Scholar]
- 44.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 45.Morrisey J. Silver stain for proteins in polyacrylamide gels: A modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981;117:307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- 46.Philip-Hollingsworth S, Hollingsworth RI, Dazzo FB. Host-range related structural features of the acidic extracellular polysaccharides of Rhizobium trifolii and Rhizobium leguminosarum. J Biol Chem. 1989;264:1461–1466. [PubMed] [Google Scholar]
- 47.Velázquez E, Mateos PF, Pedrero P, Dazzo FB, Martínez-Molina E. Attenuation of symbiotic effectiveness by Rhizobium meliloti SAF22 related to the presence of a cryptic plasmid. Appl Environ Microbiol. 1995;61:2033–2036. doi: 10.1128/aem.61.5.2033-2036.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.