Abstract
Activation of the MEK/ERK/Elk-signaling cascade is a mechanism for relaying mitogenic and stress stimuli for gene activation. MEK1 is the proximate kinase for activation of ERK1/2, and nuclear targeting of ERK1/2 is obligatory for Elk1 transcriptional activity. Human biliverdin reductase (hBVR) is a recently described Ser/Thr/Tyr kinase in the MAPK insulin/insulin-like growth factor 1 (IGF1)-signaling cascade. Using 293A cells and in vitro experiments, we detail the formation of a ternary complex of MEK/ERK/hBVR, activation of MEK1 and ERK1/2 kinase activities by hBVR, and phosphorylation of hBVR by ERK1/2. hBVR is nearly as effective as IGF1 in activating ERK; intact hBVR ATP-binding domain is necessary for Elk1 activation, whereas protein–protein interaction is the basis for hBVR activation of MEK1 and ERK. The two MAPK docking consensus sequences present in hBVR, F162GFP and K275KRILHCLGL (C- and D-box, respectively), are ERK interactive sites; interaction at each site is critical for ERK/Elk1 activation. Transfection with mutant hBVR-P165 or peptides corresponding to the C- or D-box blocked activation of ERK by IGF1. Transfection with D-box mutant hBVR prevented the activation of ERK by wild-type protein and dramatically decreased Elk1 transcriptional activity. hBVR is a nuclear transporter of ERK; experiments with hBVR nuclear export signal (NES) and nuclear localization signal (NLS) mutants demonstrated its critical role in the nuclear localization of IGF-stimulated ERK for Elk1 activation. These findings, together with observations that si-hBVR blocked activation of ERK and Elk1 by IGF1 and prevented formation of ternary complex between MEK/ERK/hBVR, define the critical role of hBVR in ERK signaling and nuclear functions of the kinase.
Keywords: Elk-1, ERK docking sites, ERK MAP kinase, heme oxygenase-1, adapter/scaffold proteins
Biliverdin reductase (BVR) is an evolutionarily conserved enzyme expressed in a broad range of tissues. It was initially characterized by its unique dual pH/cofactor reductase activity profile in the heme metabolism pathway. Recent studies have characterized the human BVR (hBVR) as a member of the rare type of dual-specificity (Ser/Thr/Tyr) kinases and an interactive kinase in the insulin/insulin-like growth factor 1 (IGF1)-signaling pathways (1). hBVR is a soluble protein, whereas most tyrosine kinases, such as the insulin receptor kinase (IRK), are membrane-associated (2). The extracellular signal-regulated kinases, ERK1/2, p38 and JNK, are the main subfamilies of MAPKs in the insulin/IGF1-signaling pathways, with activation outcomes that include cell proliferation, differentiation, apoptosis, and immune response (3, 4). The outcomes reflect altered expression of genes activated by a wide variety of stimuli, broadly categorized as growth factors and stress inducers (4). The ERK1/2 kinases are primarily growth factor-activated, whereas p38 and JNK/SAPK1 are stress-activated. Other members of the ERK family, such as ERK3, -5, and -7, are less well characterized.
Several observations suggested the potential link between hBVR and ERK for the induction of gene expression. Both stimuli categories that activate MAPK also activate hBVR—including mitogens, cytokines, free radicals, and insulin (1, 5, 6). MEK1/2 (MAPKK), the immediate upstream activator of ERK1/2, is activated by PKCζ, which plays a central role in the activation of ERK1/2 by cytokines (7). Notably, hBVR is an activator of this PKC as well as the conventional PKCβII (5, 6) and functions as an adapter/scaffold and intracellular transporter of the PKCs. Activation of PKCs and ERK is associated with the induction of AP-1 transcription factors (c-Jun, c-Fos, and ATF-2/CREB), which are downstream targets of MAPK (8). Activation of hBVR has been correlated with the aforementioned AP-1 transcription factor induction and NF-κB that acts downstream of the MAPK/PI3K/Akt-signaling cascade (9–11); the stress-activated enzymes HO-1 and iNOS are among the targets of the transcription factors (12–15). Moreover, an outcome of ERK activation is cell differentiation and proliferation; notably, the overexpression of hBVR results in a striking change in cell morphology (16).
Many of the nearly 50 potential ERK substrates identified are crucial to ERK1/2-mediated changes in the gene expression profile and require its nuclear localization (17, 18). The transcription factor Elk1 is among the nuclear targets of ERK. ERK is sequestered in the cytoplasm in complex with PEA-15, which also may function in ERK export from the nucleus (19). Because ERK1/2 lack sequences required for nuclear import or export, the nuclear targeting of the activated dimeric proteins, which is an energy-dependent process, is mediated through association with NLS- and/or NES-containing soluble shuttling carrier(s) (17–19). The identity of the nuclear import/export carrier for ERK1/2 remains unclear, although MEK1, which has a NES, and the scaffolding protein GAB1, which has a NLS but is not a kinase, have been suggested to play roles in nuclear trafficking of ERK.
Several features of hBVR led us to hypothesize that it is an ERK import/export carrier. Aside from being a kinase, hBVR has a functional NLS (6) and a leucine-rich segment, L176VSLFGELSL, which is a characteristic NES signal that functions in nuclear-export proteins (20). In addition, the secondary structure of hBVR, as predicted by that of rat BVR, is favorable for protein–protein interaction, particularly in the carboxyl domain, which contains six β-strands forming a large β-sheet (21, 22). The leucine-rich domain is located in the β-sheet. The secondary structure of hBVR further predicts a structure resembling the conserved pleckstrin homology domain, a characteristic of scaffold proteins (23, 24). Those proteins either recruit enzymes into signaling networks or assemble them in proximity to their substrates (24). Tyrosine is phosphorylated by insulin-activated IRK in two hBVR consensus SH2-binding motifs: Y198MKM and Y228LSF (1). YMXM and YϕS/Tϕ motifs provide the docking sites for SH2 domain adaptor signaling proteins such as Grb-2, the GAB1 adaptor protein (24).
The C-terminal α-helix of hBVR contains a sequence, K275KRILHCLGL, similar to the D- (δ) box/domain found in ERK1/2 and JNK substrates; it has a conserved consensus sequence, (R/K)2X2–6L/IX1L/I (12, 25). The D-box sequence does not confer substrate specificity for MAPK/ERK subfamilies by itself. A second sequence, FXFP or C-box, provides the specific binding and substrate recognition site for ERK (25). The C-box is identical to the hBVR F162GFP sequence. The presence of this sequence in the Elk subfamily of ERK substrates ensures high-affinity binding of these proteins to ERK (25). Both hBVR and Elk1 are transcription factors with overlapping target genes: Elk1 is an ETS-domain transcription factor that binds to target promoters after phosphorylation and dissociation from ERK (26), whereas hBVR is a bZip transcription factor and binds to 7/8-bp AP1 sites (10). A comparison of hBVR and Elk1 primary structures is presented in supporting information (SI) Fig. S1.
Based on the noted features and functions of hBVR, we hypothesized that it plays a critical role in ERK/Elk nuclear signaling. The study, using a combination of in vitro and in-cell approaches, has identified hBVR as the ERK activator, nuclear transporter, and regulator of the MEK/ERK/Elk-signaling cascade. Data suggest that hBVR plays a pivotal role in the regulation of gene expression in response to the activation of the MAPK pathway by extracellular stimuli.
Results
hBVR Binds to and Activates ERK1/2 and Is Phosphorylated by ERK1/2.
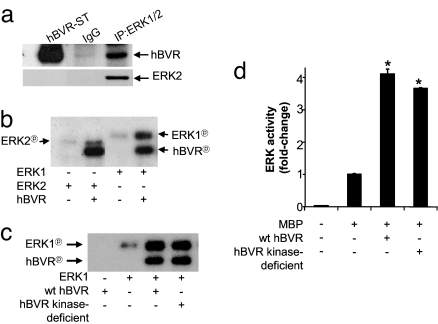
We examined the interaction of ERK and hBVR by using the immunoprecipitation (IP) assay approach. Cell lysates transfected and treated with hBVR and IGF1, respectively, were IPed with antibody to ERK1/2 and were analyzed by Western blotting for the presence of hBVR. Fig. 1a shows that hBVR, together with ERK2, was present in the IP after IGF1 treatment, but not in the IgG control. Next, whether hBVR activates ERK1/2 and whether ERKs phosphorylate hBVR were examined by using assay conditions that do not support hBVR kinase activity (1, 5, 6). ERK1/2 autophosphorylation was stimulated in the presence of hBVR, and a concurrent increase in hBVR phosphorylation was evident (Fig. 1b). An increase in ERK autophosphorylation was concentration-dependent (Fig. S2).
Fig. 1.
hBVR-ERK1/2 binding increases ERK1/2 and hBVR phosphorylation. (a) hBVR binds to ERK1/2. HEK293A cells transfected with hBVR and treated with IGF1 were lysed and IPed with polyclonal ERK1/2 antibodies or control IgG. Western blot was sequentially probed with hBVR and monoclonal ERK2 antibodies. ST, standard. (b) hBVR is a substrate and activator of ERK1 and ERK2 autophosphorylation. The kinase activities of ERK2 and ERK1 were measured with or without GST-hBVR, and phosphorylation was detected by autoradiography. (c) ERK1 activation by hBVR is independent of its kinase competency. ERK1 kinase assay was incubated in vitro with hBVR or its kinase-deficient mutant as in b. (d) ERK kinase activity is increased by hBVR. GST-ERK1 was assayed by using MBP as the substrate in the presence of hBVR or its kinase-deficient form. Experimental details are as described in Materials and Methods. *, P < 0.001 vs. no hBVR.
To establish that activation of ERK is solely the result of hBVR binding, ERK kinase activity was measured by using kinase-deficient hBVR (V11–14→A, G17→A). Nearly identical activation was observed with the mutant and wild-type (wt) hBVR as assessed by autophosphorylation (Fig. 1c) and by phosphorylation of MBP substrate (Fig. 1d). Activation of ERK by hBVR was neither an in vitro phenomenon nor cell line-specific because H9C2 rat cardiomyoblasts transfected with hBVR displayed a similar enhancement of ERK kinase activity as HEK293A cells (Fig. S3).
hBVR Forms a Ternary Complex with ERK and MEK and Activates MEK in Cells.
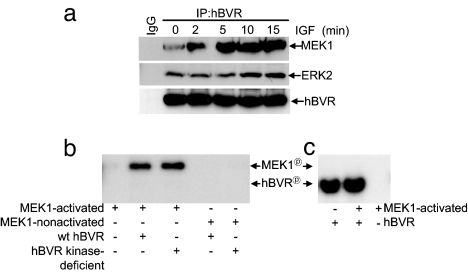
Because ERK is activated in vivo by MEK1, the above observations raised the question of whether hBVR was acting independently of MEK1 or participating in the MEK1/ERK activation complex. For this, lysates obtained from cells cotransfected with hBVR and MEK1-HA expression plasmids and treated with IGF1 were IPed with anti-hBVR. The IP was examined for the presence of MEK1 and ERK2 by Western blotting by using antibodies to HA and ERK2. IGF1 treatment of cells resulted in a rapid increase in the levels of both MEK1 and ERK2 in the IP (Fig. 2a). Next, inquiry was made as to whether hBVR activates MEK1 and vice versa. For these studies, both c-Raf-activated and nonactive MEK1 proteins were used; MEK enzymes require activation by a MEK kinase (such as c-Raf) that phosphorylates two S/T residues in MEK, after which MEK is able to target ERK at a TXY sequence in the activation loop. These studies used both wt hBVR and the kinase-deficient form of the protein. Fig. 2b shows the presence of wt hBVR or the mutant equally stimulated autophosphorylation of MEK1. MEK autophosphorylation was not detected by using nonactivated MEK1. MEK was not a substrate for hBVR under assay conditions that support hBVR autophosphorylation (Fig. 2c). Collectively, the findings of Figs. 1 and 2 indicate that hBVR activates both ERK and MEK1; however, although it is not a substrate for MEK, it is a substrate for ERK.
Fig. 2.
hBVR forms a ternary complex with ERK1/2 and MEK1. (a) ERK and MEK1 co-IP with hBVR. Cells cotransfected with hBVR and MEK1-HA expression plasmids were treated with IGF1. Western blotting and sequential probing with anti-HA, ERK2, and hBVR antibodies analyzed the IP obtained by using anti-hBVR. (b) hBVR is not a substrate for MEK1. c-Raf-activated or nonactivated MEK1 proteins were incubated with wt- or kinase-deficient hBVR, and products were visualized as in Fig. 1b. (c) hBVR does not phosphorylate MEK1. hBVR kinase activity using MEK1 as substrate was analyzed as in b.
hBVR Activates ERK/Elk1 Signaling Independent of IGF1.
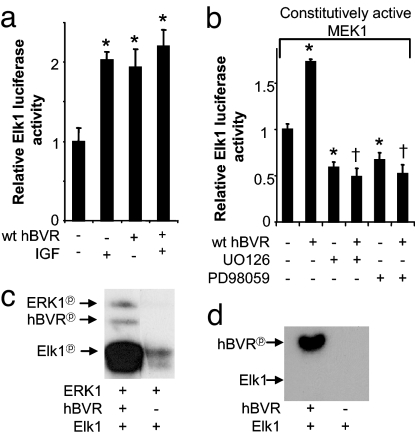
The effect of hBVR on the Elk1-dependent promoter activity was measured in cells transfected with the Elk1-luciferase reporter plasmids either cotransfected with hBVR, treated with IGF1, or both. As noted in Fig. 3a, hBVR alone significantly stimulated Elk1-dependent transcription, and the extent of activation was similar to IGF1. Elk1 stimulation was not further increased when both hBVR and IGF1 were present. Subsequently, a dominant-active MEK1 was used to define whether hBVR activates Elk1 independent of upstream activators of MEK1 in cells cotransfected with MEK1 expression plasmid and hBVR or MEK1 alone (Fig. 3b). The presence of hBVR significantly increased promoter activity, suggesting that hBVR activates Elk1 parallel or downstream of MEK. Whether hBVR acts downstream of MEK1 was further examined by treating cells with the MEK1/2 inhibitor UO126 or the specific MEK1 inhibitor PD98059 and then testing whether Elk1 inhibition could be rescued by the overexpression of hBVR. As expected, both compounds significantly inhibited Elk1-activated transcription. However, the inhibition was not overcome by hBVR, suggesting a parallel interaction of hBVR with MEK1 in enhancing Elk1 activity.
Fig. 3.
Elk1-mediated regulation of gene transcription is stimulated by hBVR-activating MEK1/ERK1 signaling. (a) hBVR induces Elk1 activation by ERK. Cells cotransfected with hBVR, in addition to the Elk1 reporter plasmids, were starved and treated with IGF1 (16 h). Lysates were assayed by dual luciferase as described in Materials and Methods. *, P < 0.01 vs. untreated. (b) hBVR enhances constitutively active MEK1-induced Elk1 activity. Cells transfected with constitutively active MEK1 were starved and treated with 10 μM inhibitors or cotransfected with hBVR. *, P < 0.01 vs. untreated; †, P < 0.01 vs. wt hBVR alone. (c) hBVR increases ERK1-dependent Elk1 phosphorylation. ERK1 kinase activity using GST-Elk1 and hBVR as substrates was analyzed as in Fig. 1b. (d) hBVR does not phosphorylate Elk1. hBVR kinase activity using Elk1 as the substrate was analyzed as in Fig. 1b.
Because hBVR activates both ERK- and Elk1-dependent transcription activity, it was essential to examine whether hBVR directly activates Elk1 or the activation of Elk1 is mediated as a result of ERK activation by hBVR. Fig. 3c shows that the inclusion of hBVR in the ERK in vitro kinase reaction containing Elk1 resulted in significant increases in phosphorylation of both ERK1 and Elk1; the extent of activation of Elk1 is striking. The increase in Elk1 phosphorylation was not the result of hBVR kinase activity (Fig. 3d). Accordingly, this finding further indicates that hBVR activates Elk1 via the MAPK pathway at the level of MEK1.
hBVR Activation of ERK Is Dependent on its C- and D-box ERK1/2 Docking Sites.
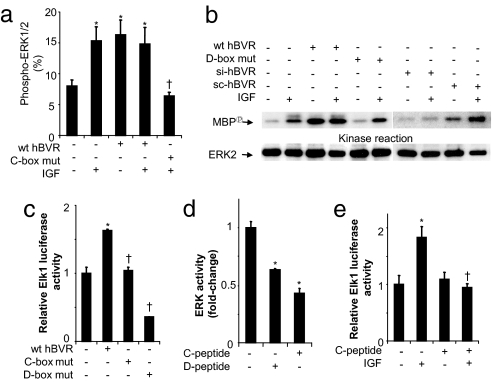
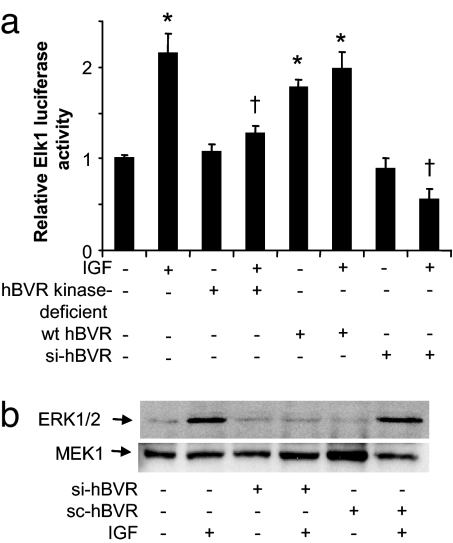
hBVR's role in ERK activation was tested by flow cytometry by using a FITC-conjugated antibody to a phospho-ERK1/2 peptide phosphorylated within the activation loop on T185 and Y187. As noted in Fig. 4a, wt hBVR overexpression caused an ≈2-fold increase in the proportion of cells stained for phospho-ERK1/2 in both control and IGF1-treated cells. Comparably, the activation of ERK by IGF1 was attenuated in the presence of the C-box mutant. Next, the relevance of the hBVR D-box to ERK activity toward MBP as a substrate was examined. In the presence of the D-box mutant hBVR, IGF1 failed to activate ERK activity (Fig. 4b). Further support for the significance of hBVR to the activation of Elk was provided by the observation that si-hBVR blocked the activation produced by wt hBVR or IGF1. That a scrambled siRNA (sc-hBVR) was ineffective in blocking ERK activation validated the observation.
Fig. 4.
hBVR C-box ERK docking site is necessary for the activation of ERK and Elk1 in the cell. (a) Flow-cytometry analysis. Cells were transfected with wt- or C-box mutant hBVR, starved, treated with IGF1, fixed, permeabilized, and treated with FITC-conjugated anti-phospho-ERK1/2 antibody. The proportion of stained cells was determined by flow cytometry. *, P < 0.01 vs. control. †, P < 0.01 vs. IGF1. (b) hBVR D-box mutant does not stimulate ERK. Cells were transfected with hBVR or D-box mutant plasmids or were infected with si-hBVR or sc-hBVR, starved, and treated with IGF1. IP obtained with anti-ERK2 was assayed for ERK activity. The blot was reprobed by using ERK2 antibody. (c) C- and D-boxes are required for hBVR-mediated induction of ELK. Cells were cotransfected with Elk1 reporters and wt hBVR, or C- or D-box mutants, and lysates were assayed for luciferase. *, P < 0.01 vs. control; †, P < 0.01 vs. wt hBVR. (d) hBVR D- and C-box peptides regulate ERK activity in vitro. Phosphorylation of MBP by ERK1 was measured in the presence of 10 μM C- or D-box peptide. *, P < 0.01 vs. control. (e) hBVR C-box peptide prevents IGF1-dependent activation of Elk1 by ERK. Cells transfected with the Elk1 reporters were treated with C-box peptide before IGF1 treatment. *, P < 0.01 vs. untreated; †, P < 0.01 vs. IGF1-treated. Experimental details are provided in Materials and Methods.
In the next experiment, the effects of hBVR C- and D-box mutants on Elk1 activation were examined. The expected activation of Elk1-dependent transcriptional activity by hBVR was observed (Fig. 4c). However, such a response was not elicited in cells transfected with C- or D-box mutant hBVR, and indeed the response markedly decreased after transfection with D-box mutant plasmid.
That the two potential ERK docking domains of hBVR are in fact the interaction sites between hBVR and ERK was confirmed by using peptides that encompassed the hBVR C-box (FGFPAFSG) and D-box (KKRILHCLGLA) in experiments that measured ERK activity and Elk1-dependent transcription. Fig. 4d shows that the kinase activity of ERK1 using MBP as the substrate decreased in the presence of either peptide. Experiments that assessed Elk1 activation in the cell by IGF1 confirmed the finding (Fig. 4e). As noted in Fig. 4e, the C-box peptide blocked the activation of ERK. Collectively, the findings with peptides suggest the likelihood of peptides binding and blocking the substrate-binding site and/or changing ERK's conformation, resulting in reduced access to substrate.
Because Elk S/TP motifs located near the C-box (Fig. S1) are the targets of ERK kinase activity, we examined whether the S211P motif in hBVR is a target of ERK phosphorylation. In addition, S230 in YLSF that flanks the hBVR NLS also was tested. This motif is one of the SH2-binding domains of hBVR and also is involved in its interaction with PKCζ. Unlike the Elk1 S/TP motifs, the unexpected finding was that the S211 mutation did not decrease hBVR phosphorylation by ERK, whereas the S230 mutation significantly decreased phosphorylation of hBVR by ERK (Fig. S4). The finding with S230 is consistent with the possibility that the YLSF motif is of significance in the insulin/IGF1/ERK activation loop. As predicted by the solved crystal structure of rat BVR (21, 22), in the human protein, S211, unlike S230, is not in a favorable position to be accessed by ERK.
hBVR Is Essential for the Activation of Elk1.
In the next series of experiments, the effect of reducing the cellular concentration of hBVR in cells infected with the siRNA construct, or transfection of cells with its kinase-deficient hBVR mutant, on Elk1-dependent transcription was examined. These experiments used control cells or cells treated with IGF1. As noted in Fig. 5a, si-hBVR and mutant hBVR blocked the IGF1-mediated increase in Elk1 activity. The si-hBVR was more effective in blocking activation than the kinase-deficient hBVR, an observation that emphasizes the role of endogenous hBVR in Elk1 activation. Results of the following experiment defined hBVR as the integral component of the ERK/MEK1 activation complex for Elk1-dependent transcriptional activity. In this experiment, native cells infected only with si-hBVR and treated with IGF1 were IPed with anti-MEK1; cells infected with sc-hBVR served as controls. As noted in Fig. 5b, the level of ERK in the IP that was obtained from IGF1-treated cells was remarkably reduced in the presence of si-hBVR. The decrease in the amount of ERK in the IP was not due to differential loading of the sample, as indicated by the observation that the MEK1 level, as assessed by Western blotting, was essentially equal in all samples. This observation was further validated by the finding that the amount of IPed ERK was indistinguishable in IGF1-treated control cells and those cells treated with IGF1 and infected with inactive sc-hBVR.
Fig. 5.
hBVR is required for MEK1/ERK/Elk1-mediated transcription. (a) The hBVR ATP-binding domain is needed for Elk1 activation. Cells were cotransfected with Elk1 reporters and wt- or kinase-deficient hBVR, or infected with si-hBVR, treated with IGF1, and assayed for luciferase reporter activity. *, P < 0.001 vs. untreated; †, P < 0.01 vs. wt. (b) si-hBVR prevents MEK1 binding to ERK. Cells infected with si-hBVR or sc-hBVR were treated with IGF1 and IPed by using MEK1 antibodies. The blot was consecutively probed with ERK1/2 and MEK1 antibodies. Experimental details are provided in Materials and Methods.
The Reductase Is a Nuclear Transporter of ERK.
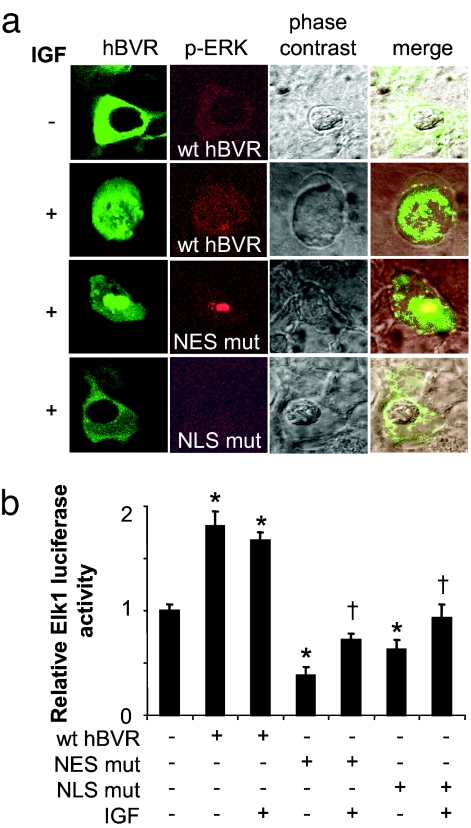
As noted above, the essential step in the activation of Elk1, and thus the transmission of stimuli in regulation of cell proliferation by the ERK1/2, is the nuclear translocation of the activated form (27, 28). A previous study showed that the hBVR NLS motif is functional in its nuclear translocation (6). We examined whether the NLS functions in transport of a complex of hBVR with another protein, ERK, into the nucleus. Also investigated was whether the consensus NES motif is functional and participates in nuclear trafficking of the ERK/hBVR complex. In the first series of experiments, confocal microscopy was used to visualize the cellular localization of hBVR and phosphorylated ERK. As depicted in Fig. 6a, in untreated cells, EGFP-hBVR and phosphorylated ERK were located primarily in the cytoplasm. IGF1 treatment of the cells resulted in the redistribution of both proteins between the nucleus and the cytoplasm. In contrast, neither the hBVR NLS mutant nor the phospho-ERK translocated to the nucleus. In cells expressing the hBVR NES mutant, it is apparent that there is significant accumulation of hBVR in the nucleus, which is accompanied by concomitant accumulation of phospho-ERK. Notably, a similar observation (i.e., accumulation of phospho-ERK and MEK1 in the nucleus) has been made for MEK1 with a disrupted NES (29). Analysis of Elk1 reporter activity (Fig. 6b) in cells transfected with wt hBVR or its NLS or NES mutants agrees with the confocal microscopy data and confirms the relevance and functionality of the hBVR targeting sequences to ERK nuclear localization and activity. To confirm that hBVR translocates into the nucleus, a second method of analysis was used by comparing the level of hBVR in the cytosol and nucleus in cells transfected with EGFP-hBVR plasmid and treated with IGF1. Measurement of hBVR signal in the enriched nuclear fraction and the cytosol confirmed the confocal microscopy data (Fig. S5).
Fig. 6.
hBVR NLS and NES motifs are involved in the transport of ERK. (a) Localization of hBVR and pERK in IGF1-treated cells. HEK293A cells were transfected with pEGFP-hBVR or its mutants and then treated with IGF1 15 min before fixation and treatment with the pERK1/2T185Y187 primary and Rhodamine-conjugated secondary antibodies. Confocal microscopy was performed as described previously (5, 6). (b) hBVR NES and NLS are required for the activation of Elk1 by IGF1. Cells cotransfected with Elk1 reporters and wt hBVR or mutants were starved before treatment with IGF1. After 18 h, luciferase was assayed as described in Materials and Methods. *, P < 0.01 vs. untreated cells; †, P < 0.001 vs. wt.
Discussion
The primary structure of hBVR possesses an unprecedented array of consensus regulatory motifs, many of which are functional and account for the diversity of its roles in insulin/IGF1/MAPK signal-transduction pathways (1, 5, 6). Relying on the endogenous ERK, in this study, by disrupting consensus sequences of NES, NLS, C- and D-box ERK-binding domains, ATP-binding pocket, or ablating hBVR, we have assembled reliable evidence for two integrated mechanisms by which hBVR regulates ERK nuclear activity: One solely involves protein–protein interaction, whereas the other function requires both its kinase activity and binding.
We show that kinase-competent hBVR exerts control over the nuclear activity of the signaling pathway by functioning in the nuclear import/export of ERK. In doing so, hBVR alone can mimic functions that have been ascribed to several proteins. In this capacity, hBVR links the cytoplasmic events to nuclear processes that result in the induction of genes involved in cell proliferation and differentiation. Noteworthy, too, is the reductase activity of the enzyme that provides the sole source of bilirubin while rescuing the activity of enzymes and kinases that are inhibited by biliverdin, including that of HO-1. Bilirubin is an inhibitor of inflammatory responses and mitogen-induced reactive oxygen species (ROS) production; activation of ERK1/2 by ROS is among the cell-signaling events that lead to the induction of the cytoprotective gene HO-1 (14) (see SI Text).
hBVR, like the Drosophila KSR protein (kinase suppressor of Ras) (30) and the MEK partner 1 protein (MP1) (31), is an activator of MEK and, as suggested by our data, a partner in linking ERK to activation by MEK. hBVR functions upstream of ERK within the MEK/ERK module and interacts with both MEK and ERK to form a complex as is indicated by the observation that the cell lysate IPed with antibody to hBVR contained hBVR, MEK, and ERK; in the complex, hBVR functions as a scaffold. Although it is neither a substrate (Fig. 2b), a substitute (Fig. 3b), nor a kinase for MEK (Fig. 2c), MEK activation of ERK is blocked by inhibitory si-hBVR (Fig. 4b). As a scaffold, it appears that hBVR, by binding to the two proteins, places ERK in a position that allows its activation by MEK. This is not intended to suggest that the assembly is made only of MEK/ERK/hBVR. Furthermore, because inhibitors of MEK also inhibit Elk1-dependent transcription, the binding places hBVR parallel to MEK in the MAPK-signaling network.
In light of the present observations, the question may be raised as to why involvement of hBVR in the nucleus import and export of ERK was not uncovered in the course of previous studies. This, we suggest, reflects the fact that, although hBVR is ubiquitously present in tissues, it went undetected because of the unavailability of antibodies to hBVR for studies that primarily rely on the IP approach.
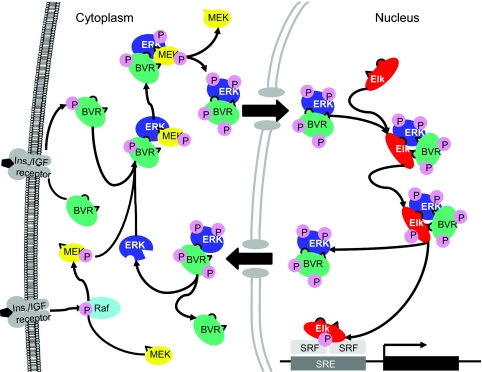
Downstream of ERK, hBVR shares certain structural similarities with Elk1 (Fig. S1) by having two ERK-binding sites that allow high-affinity binding between ERK and its substrates. These are the D-box docking site, which is common to all MAPK substrates, and the second substrate binding site, FXFP, which is specific to ERK1/2 and is exposed to activated ERK (12, 25). The two sites function additively, with the FXFP site imparting high-affinity binding to ERK. Our data suggest that the common denominator for the activity of hBVR in the cytosol and the nucleus is its high-affinity binding to ERK and the ability to exchange a second binding partner depending on cellular location. In the cytosol, hBVR is a component of a ternary complex with MEK/ERK; upon activation, ERK is released from MEK with the high-affinity hBVR/ERK complex entering the nucleus where a new ternary module of hBVR/ERK/Elk is formed. The proposal also is consistent with the following notion: There is no FXFP ERK docking site in MEK1, which would limit docking to the low-affinity D-domain of MEK. Upon ERK phosphorylation, the MEK/ERK complex dissociates, with MEK staying in the cytoplasm (19). The proposed mechanism is supported by the following data: (i) IP experiments demonstrate that hBVR antibodies coprecipitate the antigen together with MEK and ERK; (ii) in the presence of hBVR, there is a striking increase in the phosphorylation of Elk and activation of Elk1-dependent transcription; and (iii) si-hBVR hinders Elk1-dependent transcription activity. A schematic presentation of the findings is provided in Fig. 7.
Fig. 7.
Proposed functions of hBVR in the MAPK-signaling pathway. The C- and D-boxes of hBVR, MEK, and Elk1 are indicated by the rounded and triangular protuberances, respectively.
The present findings, together with previous data demonstrating the activation of PKCβII and PKCζ through protein–protein interaction with hBVR, independent of hBVR kinase activity (5, 6), suggest that physical interaction among hBVR and components of the signaling network is an important mechanism for the integration and transmission of extracellular signals to stimulate transcriptional activity. Here we found that transfection with hBVR plasmid was as effective as IGF1 in stimulating ERK activation of Elk1 reporter activity (Fig. 3 a and b). That an inactive kinase functions as a scaffold molecule independent of kinase activity is not unprecedented. For example, KSR1 also possesses kinase activity, which is not relevant to its role as a MEK/ERK scaffold protein (30). Accordingly, we propose that the scaffolding and kinase functions may be more or less important depending on the cellular localization, but that both activities are essential for the full range of signaling activity.
In conclusion, the present study reveals the multidimensional input of hBVR in the regulation of extracellular-regulated MAPK signaling that encompasses functions attributed to several proteins and, as such, points to the likelihood of hBVR being a central component of signal transduction in mammalian cells.
Materials and Methods
Materials.
hBVR-based myristoylated peptides K275KRILHCLGLA (D-box) and F162GFPAFSG (C-box) were synthesized by EZBiolab. MBP, MEK1 proteins, and antibody against MEK1 were obtained from Upstate Biotechnology. Sources of ERK1 and ERK2 active proteins and antibodies were EMD and BD Biosciences, respectively, while monoclonal anti-ERK2, anti-phospho-ERK1/2, and Elk1 fusion protein (amino acids 307–428) were from Santa Cruz Biotechnology. Polyclonal anti-human BVR antibodies were generated as before (9). HA antibody and A/G agarose beads were from Santa Cruz Biotechnology. UO126 and PD98059 were obtained from Calbiochem.
Plasmids.
Target amino acid(s) of a wt cDNA clone were replaced with alanine. For the kinase-defective hBVR, V11–14 and G17 were replaced; for the hBVR NLS mutant, A222IGSTG was replaced with G222LKRNR; and for the NES mutant, L176VSLFGELSL was changed to A176VSVFGEVSA. The D-box mutant has I278LHCLGL changed to A278AHCAGA; the C-box mutant has P165 replaced. Mutations were made in pGEX-4T2 for Escherichia coli expression or in pcDNA3 or EGFP fusions for expression in mammalian cells. The plasmid pFC-MEK1 (Stratagene) encodes the constitutively active Δ32–61, S218E, S222E mutant. Expression plasmids for ERK1/2 and dominant-negative ERK1/2 were generous gifts of Melanie H. Cobb (Southwest Washington Medical Center, Dallas, TX).
Cell Culture, Transfection, and Treatment.
HEK293A cells were grown in DMEM (Invitrogen) containing 10% FBS to 70% confluency. Cells were transfected with plasmids, as appropriate, by using Transfectin (Bio-Rad). pSuper-si-hBVR or sc-hBVR were prepared as before (10). Cells were infected with four plaque-forming units per cell. Then 20 ng/ml IGF1 (15 min) or peptides were added to cells starved for 18 h in medium containing 0.1% FBS. When used together, peptide was added 2 h before IGF1.
IP.
Cells were transfected, starved, treated with IGF1 or vehicle, and lysed as described above. Lysates, prepared as in ref. 5, containing equal amounts of protein were IPed with the appropriate antibodies, followed by the addition of A/G agarose. IPs were separated by SDS/PAGE, transferred to a PVDF membrane, and detected by using enhanced chemiluminescence (Amersham Pharmacia Biotech).
ERK, MEK1, and hBVR Kinase Assays in Vitro.
Briefly, 5 ng of active ERK1 or 2 or 6 ng of MEK1 active was used in the kinase assay. The reaction was initiated by the addition of 100 μM [γ-32P]ATP and terminated by the addition of Laemmli buffer, followed by SDS/PAGE, transfer to PVDF membrane, and autoradiography. Kinase activity of hBVR was assayed as described (1, 5, 6). Alternatively, termination was achieved by the addition of 1 volume of 10% phosphoric acid, and incorporation of 32P was measured by binding to Whatman P81 filters (1).
Kinase Assay for Endogenous ERK.
A/G-agarose-bound ERK2 IPs (using anti-ERK2 antibody, which cross-reacts weakly with ERK1) were prepared from HEK293A cell lysates and then washed with lysis buffer containing 200 mM LiCl, and kinase assay buffer. The ERK kinase activity was analyzed as above.
Flow Cytometry.
Cells were detached with 2 mM EDTA, suspended in serum-free DMEM, treated with IGF1, if appropriate, and fixed with 1% paraformaldehyde in PBS. Cells permeabilized in ice-cold methanol were blocked with 0.1% saponin, 2% FBS in PBS for 30 min, and treated with FITC-conjugated antibody to ERK1/2 for an additional 30 min. Cells were sequentially washed with the blocking solution and cold PBS and then sorted by flow cytometry.
Elk1-Luciferase Reporter Assays.
Cells were cotransfected with pFA2-Elk1, pRF-Luc, and pRL-TK (PathDetect Elk1 transreporting system; Stratagene) together with other plasmids, as suggested by the manufacturer. Cells were lysed and promoter/transcription activity was measured by using a dual-luciferase assay system (Promega).
Confocal Microscopy.
Cells grown in a chamber slide system (Nalge Nunc) were transfected with pEGFP-hBVR or mutants (10). After 18 h starvation, they were treated with 20 ng/ml IGF1 or vehicle for 15 min and processed as before (5, 6). Mouse anti-ERK antibody was used at a 1:500 dilution. PBS-washed cells were treated with Rhodamine-red-X-conjugated donkey anti-mouse IgG antibodies for 30 min. Images were obtained by using a Leica TCS SP model DMRE confocal microscope.
Data Analysis.
Each experiment was repeated at least three times. Data are shown as mean ± SD. Statistical analysis was performed by Student's t test using GraphPad prism software.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grants ES 04066 and ES012187.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800750105/DCSupplemental.
References
- 1.Lerner-Marmarosh N, et al. Human BVR: A member of the insulin receptor substrate family with serine/threonine/tyrosine kinase activity. Proc Natl Acad Sci USA. 2005;102:7109–7114. doi: 10.1073/pnas.0502173102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter T. The role of tyrosine phosphorylation in cell growth and disease. Harvey Lect. 1998;94:81–119. [PubMed] [Google Scholar]
- 3.Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: A family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev. 2004;68:320–344. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karin M. Inflammation-activated protein kinases as targets for drug development. Proc Am Thorac Soc. 2005;2:386–390. 394–395. doi: 10.1513/pats.200504-034SR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lerner-Marmarosh N, Miralem T, Gibbs PEM, Maines MD. Regulation of TNF-α-activated PKC-ζ signaling by the hBVR: Identification of activating and inhibitory domains of the reductase. FASEB J. 2007;21:3949–3962. doi: 10.1096/fj.07-8544com. [DOI] [PubMed] [Google Scholar]
- 6.Maines MD, Miralem T, Lerner-Marmarosh N, Shen J, Gibbs PEM. Human BVR, a previously unknown activator of PKC βII. J Biol Chem. 2007;282:8110–8122. doi: 10.1074/jbc.M513427200. [DOI] [PubMed] [Google Scholar]
- 7.Monick MM, Carter AB, Flaherty DM, Peterson MW, Hunninghake GW. Protein kinase C ζ plays a central role in activation of the p42/44 mitogen-activated protein kinase by endotoxin in alveolar macrophages. J Immunol. 2000;165:4632–4639. doi: 10.4049/jimmunol.165.8.4632. [DOI] [PubMed] [Google Scholar]
- 8.Soh JW, Lee EH, Prywes R, Weinstein IB. Novel roles of specific isoforms of PKC in activation of the c-fos serum response element. Mol Cell Biol. 1999;19:1313–1324. doi: 10.1128/mcb.19.2.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kravets A, Hu Z, Miralem T, Torno MD, Maines MD. Biliverdin reductase: A novel regulator for induction of ATF-2 and HO-1. J Biol Chem. 2004;279:19916–19923. doi: 10.1074/jbc.M314251200. [DOI] [PubMed] [Google Scholar]
- 10.Miralem T, Hu Z, Torno MD, Lelli KM, Maines MD. Small interference RNA-mediated gene silencing of hBVR, but not that of HO-1, attenuates arsenite-mediated induction of the oxygenase and increases apoptosis in 293A kidney cells. J Biol Chem. 2005;280:17084–17092. doi: 10.1074/jbc.M413121200. [DOI] [PubMed] [Google Scholar]
- 11.Gibbs PEM, Maines MD. Biliverdin inhibits activation of NF-κB: Reversal of inhibition by hBVR. Int J Cancer. 2007;121:2567–2574. doi: 10.1002/ijc.22978. [DOI] [PubMed] [Google Scholar]
- 12.Minden A, Karin M. Regulation and function of the JNK subgroup of MAP kinases. Biochim Biophys Acta. 1997;1333:F85–F104. doi: 10.1016/s0304-419x(97)00018-8. [DOI] [PubMed] [Google Scholar]
- 13.Erusalimsky JD, Moncada S. Nitric oxide and mitochondrial signaling: From physiology to pathophysiology. Arterioscler Thromb Vasc Biol. 2007;27:2524–2531. doi: 10.1161/ATVBAHA.107.151167. [DOI] [PubMed] [Google Scholar]
- 14.Ryter SW, Alam J, Choi AM. HO-1/CO: From basic science to therapeutic applications. Physiol Rev. 2006;86:583–650. doi: 10.1152/physrev.00011.2005. [DOI] [PubMed] [Google Scholar]
- 15.Immenschuh S, Fahimi HD, Baumgart-Vogt E. Complementary regulation of HO-1 and peroxiredoxin I gene expression by oxidative stress in the liver. Cell Mol Biol. 2005;51:471–477. [PubMed] [Google Scholar]
- 16.Maines MD. New insights into BVR functions: Linking heme metabolism to cell signaling. Physiology. 2005;20:382–389. doi: 10.1152/physiol.00029.2005. [DOI] [PubMed] [Google Scholar]
- 17.Ranganathan A, Yazicioglu MN, Cobb MH. The nuclear localization of ERK2 occurs by mechanisms both independent of and dependent on energy. J Biol Chem. 2006;281:15645–15652. doi: 10.1074/jbc.M513866200. [DOI] [PubMed] [Google Scholar]
- 18.Yazicioglu MN, et al. Mutations in ERK2 binding sites affect nuclear entry. J Biol Chem. 2007;282:28759–28767. doi: 10.1074/jbc.M703460200. [DOI] [PubMed] [Google Scholar]
- 19.Formstecher E, et al. PEA-15 mediates cytoplasmic sequestration of ERK MAP kinase. Dev Cell. 2001;1:239–250. doi: 10.1016/s1534-5807(01)00035-1. [DOI] [PubMed] [Google Scholar]
- 20.Kutay U, Guttinger S. Leucine-rich nuclear-export signals: Born to be weak. Trends Cell Biol. 2005;15:121–124. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Kikuchi A, et al. Crystal structure of rat BVR. Nat Struct Biol. 2001;8:221–225. doi: 10.1038/84955. [DOI] [PubMed] [Google Scholar]
- 22.Whitby FG, Phillips JD, Hill CP, McCoubrey W, Maines MD. Crystal structure of a biliverdin IXα reductase enzyme-cofactor complex. J Mol Biol. 2002;319:1199–1210. doi: 10.1016/S0022-2836(02)00383-2. [DOI] [PubMed] [Google Scholar]
- 23.Blomberg N, Baraldi E, Nilges M, Saraste M. The PH superfold: A structural scaffold for multiple functions. Trends Biochem Sci. 1999;24:441–445. doi: 10.1016/s0968-0004(99)01472-3. [DOI] [PubMed] [Google Scholar]
- 24.Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs D, Glossip D, Xing H, Muslin AJ, Kornfeld K. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 1999;13:163–175. [PMC free article] [PubMed] [Google Scholar]
- 26.Kasza A, et al. The ETS domain transcription factor Elk-1 regulates the expression of its partner protein, SRF. J Biol Chem. 2005;280:1149–1155. doi: 10.1074/jbc.M411161200. [DOI] [PubMed] [Google Scholar]
- 27.Treisman R. Regulation of transcription by MAPK cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 28.Whitmarsh AJ, Davis RJ. Transcription factor AP-1 regulation by MAPK signal transduction pathways. J Mol Med. 1996;74:589–607. doi: 10.1007/s001090050063. [DOI] [PubMed] [Google Scholar]
- 29.Fukuda M, Gotoh I, Adachi M, Gotoh Y, Nishida E. A novel regulatory mechanism in the MAP kinase cascade. Role of nuclear export signal of MAP kinase kinase. J Biol Chem. 1997;272:32642–32648. doi: 10.1074/jbc.272.51.32642. [DOI] [PubMed] [Google Scholar]
- 30.Roy F, Laberge G, Douziech M, Ferland-McCollough D, Therrien M. KSR is a scaffold required for activation of the ERK/MAPK module. Genes Dev. 2002;16:427–438. doi: 10.1101/gad.962902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaeffer HJ, et al. MP1: A MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science. 1998;281:1668–1671. doi: 10.1126/science.281.5383.1668. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.