Abstract
Genes encoding elongation factor-like (EFL) proteins, which show high similarity to elongation factor-1α (EF-1α), have been found in phylogenetically distantly related eukaryotes. The sporadic distribution of “EFL-containing” lineages within “EF-1α-containing” lineages indirectly, but strongly, suggests lateral gene transfer as the principal driving force in EFL evolution. However, one of the most critical aspects in the above hypothesis, the donor lineages in any putative cases of lateral EFL gene transfer, remained unclear. In this study, we provide direct evidence for lateral transfer of an EFL gene through the analyses of 10 diatom EFL genes. All diatom EFL homologues tightly clustered in phylogenetic analyses, suggesting acquisition of the exogenous EFL gene early in diatom evolution. Our survey additionally identified Thalassiosira pseudonana as a eukaryote bearing EF-1α and EFL genes and secondary EFL gene loss in Phaeodactylum tricornutum, the complete genome of which encodes only the EF-1α gene. Most importantly, the EFL phylogeny recovered a robust grouping of homologues from diatoms, the cercozoan Bigelowiella natans, and the foraminifer Planoglabratella opecularis, with the diatoms nested within the Bigelowiella plus Planoglabratella (Rhizaria) grouping. The particular relationships recovered are further consistent with two characteristic sequence motifs. The best explanation of our data analyses is an EFL gene transfer from a foraminifer to a diatom, the first case in which the donor–recipient relationship was clarified. Finally, based on a reverse transcriptase quantitative PCR assay and the genome information of Thalassiosira and Phaeodactylum, we propose the loss of elongation factor function in Thalassiosira EF-1α.
Keywords: EF-1α, EFL, eukaryotic phylogeny
Translation elongation factor 1α (EF-1α) and its eubacterial orthologue, EF-Tu, have been considered as indispensable proteins involved in the elongation step of protein synthesis (1, 2). Because of the critical tasks of translation elongation factors, it is widely believed that EF-1α/EF-Tu genes have been vertically inherited from the last universal common ancestor (3–5), and the gene products are ubiquitous in all extant cells. However, large-scale sequence data from phylogenetically diverged organisms started unveiling cases that clearly violate the above preconception about EF-1α/EF-Tu evolution. First, multiple cases of lateral EF-1α/EF-Tu gene transfer are described in refs. 6–8. Second, the absolute ubiquity of EF-1α has also been called into question. For instance, no EF-1α gene has been identified in the complete nuclear genome of the green alga Chlamydomonas reinhardtii (genome.jgi-psf.org). Instead, this organism possesses the gene encoding an elongation factor-like (EFL) protein that bears sequence similarity to, but is clearly divergent from, EF-1α (9). An in silico analysis of functional divergence between EF-1α and EFL suggested that EFL possesses at least a primary EF-1α function, such as the catalysis of nascent peptide elongation (9).
Although EF-1α and EFL likely share the same cellular function, the postulated evolutionary mode of transmission for EFL genes is significantly different from that for EF-1α genes, which mainly comprises vertical gene inheritance. Recent surveys of EFL genes revealed that (i) “EFL-containing” lineages are scattered amongst “EF-1α-containing” lineages and (ii) EF-1α and EFL genes are mutually exclusively distributed amongst eukaryotes (9–12). Such EF-1α/EFL distribution can be achieved by the lateral gene transfer (LGT) scenario assumes that all eukaryotes primarily lacked EFL genes, and the EFL genes were separately spread into distantly related lineages via LGT. The exogenous EFL likely took over EF-1α functions, and the endogenous EF-1α genes eventually disappeared from the genomes. Alternatively, the “gene-loss” scenario, in which assumes that eukaryotes originally possessed both EF-1α and EFL genes, is also possible. After divergence of major eukaryotic groups, the mosaic EF-1α/EFL distribution could have been created by multiple independent EFL (or EF-1α) gene-loss events. Although the current data cannot definitely distinguish the evolutionary mode of EFL genes, the LGT scenario is generally preferred over the alternative scenario based on a parsimony-based argument. Because EFL-containing lineages are currently minor amongst eukaryotes, the number of events required for the LGT scenario is equal to or smaller than the number of EFL-containing lineages. However, in the gene-loss scenario, all EF-1α-containing lineages—presumably the majority of eukaryotes—would have had to experience EFL gene loss. Thus, the number of events required in this scenario would be much larger than that required in the LGT scenario. Nevertheless, the LGT scenario for EFL evolution currently depends solely on the observed mosaic EF-1α/EFL distribution (9–12). For any putative cases of lateral EFL gene transfer, phylogenetic analyses always failed to unveil the donor lineages (9–12). To achieve better understanding of EFL evolution, information regarding the donor lineages of EFL genes would be indispensable.
We here conducted a survey of EF-1α/EFL genes in 11 diatom species. Although no EFL gene has been officially reported for diatoms, we sequenced and/or identified 10 diatom EFL genes. Our data analyses indicated that an EFL gene was established early in diatom evolution and that the pennate diatom Phaeodactylum tricornutum secondarily lost. Of utmost interest is the origin of diatom EFL genes. An EFL phylogeny recovered a strongly supported clade of the EFL homologues from the cercozoan Bigelowiella natans, the foraminifer Planoglabratella opecularis, and diatoms, with the diatoms nested inside a “Rhizarian” grouping. Characteristic sequence motifs in the EFL alignment are congruous with this relationship, arguing against phylogenetic artifact. Thus, both the tree topology and sequence features suggest that an EFL gene was most likely transferred from a foraminifer to a diatom. To our knowledge, this is the first case whereby the donor of an EFL gene is clear, making it significant for a deeper understanding of EF-1α/EFL evolution. We also discussed a potential loss of elongation factor (EF) functions in EF-1α of Thalassiosira, an organism using both EF-1α and EFL.
Results and Discussion
EFL Genes in Diatoms.
Before this study, no EFL gene has been officially identified in diatoms; only EF-1α genes of the centric diatom Thalassiosira and the pennate diatom Phaeodactylum have been reported (9, 13). Nevertheless, we identified EFL sequences in public databases. In the completed genome of Thalassiosira, an EFL gene remained unannotated (JGI assigned name, TPS_42616). EFL transcripts were also detected in expressed sequence tag (EST) data of the pennate diatom Fragilariopsis cylindrus (GenBank accession nos. EL737687 and DR026671). These sequences showed an intimate affinity to each other in a preliminary EFL phylogeny (data not shown, but see Fig. 1A).
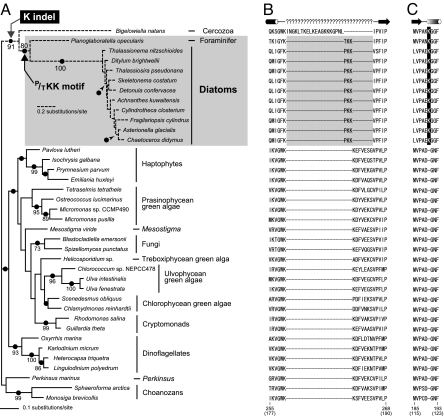
Fig. 1.
Phylogenetic relationship among EFL homologues. (A) Maximum-likelihood EFL phylogeny estimated by IQPNNI. Only bootstrap values ≥70% are indicated. The results from PhyML and MrBayes analyses are not shown, because those estimates were not significantly different from that from IQPNNI. The nodes supported by Bayesian posterior probabilities ≥ 0.95 are highlighted by dots. (B) The P/TKK motif exclusively shared among the diatom and Planoglabratella EFL homologues. Gaps are represented by dashes. Possible secondary structures deduced from a yeast EF-1α tertiary structure (PDB ID code 1IJF) are shown above the alignment. Helix and sheet are shown by cylinder and arrow, respectively. Because the secondary structure of the P/TKK motif (and the corresponding region in other EFL homologues) is unclear, question marks are inserted. Numbers below the alignment are the amino acid positions in Tharassiosira pseudonana EFL, and those in parentheses are the corresponding positions in yeast EF-1α. (C) K indel exclusively shared among the diatom, Planoglabratella, and Bigelowiella EFL homologues. Details are as described in B.
To understand more precise distribution of EFL genes in diatoms, we conducted a reverse transcriptase (RT) PCR-based survey on eight diatom species—four centric diatoms, Chaetoceros dydimus, Detonula confervenea, Ditylum brightwellii, and Skeletonema costatum, and four pennate diatoms, Asterionella glacialis, Cylindrotheca closterium, Achnanthes kuwaitensis, and Thalassionema nitzchioides (Table 1). The initial survey was conducted by using sets of degenerate PCR primers designed for amplifying short DNA fragments encoding the N termini of EF-1α and EF-1α-related proteins (i.e., EFL, eukaryotic release factor 3, hsp70 subfamily B suppressor 1). Subsequent cloning and sequencing of the amplified fragments revealed that two or more than two EF-1α/EF-1α-related genes were amplified by a single PCR for all species examined except Ditylum [supporting information (SI) Table S1]. Of note, we identified EFL genes from all of the eight species experimentally surveyed (Table 1). Nearly the entire coding regions of the eight EFL genes were then determined by 3′ rapid amplification of cDNA ends (RACE) experiments. A phylogenetic analysis of an EFL dataset recovered all diatom homologues as a robust monophyletic clade [bootstrap (BP) value = 100%; Fig. 1A], indicating that an EFL gene resided in the ancestral diatom genome before the separation of centric and pennate diatoms.
Table 1.
EF-1α and EFL genes in diatoms
| Order | Species | EFL | EF-1α |
|---|---|---|---|
| Centrales | Chaetoceros dydimus | + | N.D. |
| Detonula confervenea | + | +? | |
| Ditylum brightwellii | + | N.D. | |
| Skeletonema costatum | + | N.D. | |
| Thalassiosira pseudonana | +; 74.9 (10.9) | +; 0.192 (0.114) | |
| Pennales | Asterionella glacialis | + | +? |
| Achnanthes kuwaitensis | + | +? | |
| Cylindrotheca closterium | + | N.D. | |
| Fragiraliopsis cylindrus | + | N.D. | |
| Phaeodactylum tricornutum | − | +; 6.60 (0.780) | |
| Thalassionema nitzchioides | + | +? |
+, present; −, absent; N.D., not detected in the initial survey; +?, the initial survey amplified EF-1α gene fragment, but the rest of the gene could not be amplified by a 3′ RACE experiment. The relative copy numbers of EF-1α and EFL trascripts in Thalassiosira and that of an EF-1α transcript in Phaeodactylum are given, and standard deviations (SD) are given in parentheses.
The initial RT-PCR survey also amplified EF-1α sequences from four of the eight species (Table 1; see also Table S1). No EF-1α gene fragment was amplified by genomic DNA-based PCR experiments (data not shown). Unfortunately, we are uncertain whether these sequences were truly from diatoms, because the rest of EF-1α coding region was not obtained by subsequent RACE experiments (data not shown). In an EF-1α phylogeny, the Thalassiosira and Phaeodactylum homologues formed a monophyletic clade and further grouped with the homologues of other stramenopiles, although the particular tree topology was not strongly supported (Fig. S1).
Analyses of diatom EFL data (Fig. 1A) strongly suggested the vertical inheritance of EFL genes from the ancestor of centric and pennate diatoms. In light of this conclusion, the nuclear genome of Phaeodactylum, wherein no EFL gene is encoded, is intriguing. This pennate diatom species most likely experienced secondary EFL gene loss after divergence of the extant pennate species that branched within the centric species in molecular phylogenies of diatoms (14–16). These data display that parallel gene loss events have also shaped the EF-1α/EFL distribution in diatoms. Similar to what we observed in diatoms, the sporadic EFL distribution on a small taxonomic scale (e.g., choanoflagellates; see ref 9) is likely generated by gene loss events. Thus, gene-loss and LGT events are not mutually exclusive in EFL evolution as a whole.
Our EFL survey in diatoms also demonstrates the general importance of taxonomic sampling. Even if the complete genome is available, information without evolutionary context is not sufficient to judge whether the majority of current “EF-1α-containing” eukaryotes used to possess EFL genes in the past. In view of the incomplete EF-1α/EFL gene sampling among eukaryotes, the species that experienced secondary EFL gene loss might be more common in eukaryotes than we currently recognize.
Foraminiferan Origin of Diatom EFL Genes.
LGT has been highlighted as the principal driving force in EFL evolution, but published works (9–12) failed to clarify the donor lineages in putative cases of lateral EFL gene transfer. In sharp contrast, we successfully identified the donor lineage of the diatom EFL genes by combination of the standard phylogenetic analysis and the close examination of sequence motifs (see below).
The EFL phylogenetic analyses tightly connected the diatom clade to the homologue from the foraminifer Planoglabratella (BP = 80%; Fig. 1A). Furthermore, these homologues share three nearly identical residues in the region (the P/TKK motif; Fig. 1B), corresponding to the region between helix8 and sheet6 in a yeast EF-1α tertiary structure [Protein Data bank (PDB) ID code 1IJF; this region contains a short helix9 in the yeast structure, but the corresponding secondary structure in EFL is unclear]. The particular region in other EFL homologues was, however, occupied by seven relatively conserved residues (only the Bigelowiella homologue possesses 20 residues; Fig. 1B). Although the diatom and Planoglabratella homologues were long branches in the EFL phylogeny, the union of the two homologues bearing the P/TKK motif is unlikely a systematic artifact in tree reconstruction. Thus, we conclude that these homologues share recent ancestry excluding all others.
An additional sequence signature is also informative for resolving the origin of diatom EFL genes. Robust monophyly of Bigelowiella, Planoglabratella, and diatom EFL homologues was reconstructed (BP = 91%; Fig. 1A), and a single Lys insertion/deletion (K indel) appeared to be shared exclusively among these EFL homologues (Fig. 1C; the unique Lys residue corresponds to the first residue comprise of helix5 in yeast EF-1α). Thus, the K indel reinforces the monophyly of Bigelowiella, Planoglabratella, and diatom homologues reconstructed in the EFL phylogeny, whereas these homologues are divergent (Fig. 1A).
The monophyly of Bigelowiella, Planoglabratella, and diatom EFL homologues and the specific Planoglabratella–diatom affinity are strongly supported by both phylogenetic analysis and sequence signatures (Figs. 1 A–C). Nevertheless, the above relationship inferred from the EFL dataset is incongruent with the organismal relationships widely accepted to date. Recent phylogenetic analyses suggest close (organismal) relationship between Cercozoa (e.g., Bigelowiella) and Foraminifera (e.g., Planoglabratella) (17–19), consistent with the conception of the supergroup Rhizaria (20). However, diatoms are a subgroup of stramenopiles and do not nest inside Rhizaria (17–19). Thus, the most likely scenario for the origin of diatom EFL genes that can reconcile the discrepancy between the organismal and EFL phylogenies is as follows: (i) Bigelowiella and Planoglabratella vertically inherited an EFL gene with the K indel (Fig. 1C) from a common rhizarian ancestor. (ii) After the split of Cercozoa and Foraminifera, the foraminiferan homologues acquired the P/TKK motif (Fig. 1B). (iii) A foraminiferan EFL gene was then laterally transferred to a diatom. The tight diatom clade recovered (Fig. 1A) suggests that the EFL gene transfer took place early in diatom evolution. The foraminifer–diatom EFL gene transfer identified in this study is the first example directly supported by a series of solid phylogenetic evidence (Figs. 1 A–C). The EFL gene exchange postulated above is consistent with the biological interaction between the two protist groups in the marine environment. Foraminifers are known to host diverged eukaryotic algae including diatoms as endosymbionts (21, 22). A similar endosymbiotic foraminifer–diatom interaction in the past marine environment may have promoted the exchange of an EFL gene.
The alternative scenario invoking only the vertical transfer (and independent loss) of EFL genes cannot produce the tree topology in which diatoms nested in the rhizarian clade (Fig. 1A), because diatoms are not part of the supergroup Rhizaria (18–20). However, the direction of LGT will need to be further examined. The relationship among Bigelowiella, Planoglabratella, and diatoms recovered in the current phylogenetic analysis (Fig. 1A) is the sole evidence for the direction of this EFL gene transfer. It is generally known that phylogenetic estimates can be significantly affected by taxon (sequence) sampling. Thus, the particular relationship should be confirmed by EFL phylogenies based on datasets including more foraminiferan and cercozoan homologues than those considered in this study. It is also important to confirm that the sequence motifs (Figs. 1B and 1C) are present in other rhizarian EFL homologues.
Partial Loss of Original Functions in Thalassiosira EF-1α.
We can identify two types of diatom species with respect to the presence or absence of EFL genes: (i) “Dual-type” species, which retain EF-1α and EFL genes (e.g., Thalassiosira), and (ii) “ΔEFL-type” species, which possess only EF-1α gene (e.g., Phaeodactylum). In ΔEFL-type cells, the EF-1α may be a multifunctional protein, catalyzing the principal functions as the translation factor, and various auxiliary functions (e.g., interaction with cytoskeletal proteins and proteosomes) (1, 23, 24). However, we have no idea how EF-1α and EFL, for which the cellular functions (particularly those in peptide elongation) likely overlap, work in dual-type cells.
To obtain the first insight into the functions of EF-1α and EFL genes in dual-type cells, we investigated the transcriptional activities of the two genes in Thalassiosira by using a RT real-time PCR assay. Under laboratory culture conditions, the relative copy number of EFL transcripts appeared to be ≈75-fold higher than that of α-tubulin transcripts used as the standard (Table 1). By contrast, the EF-1α gene was expressed ≈5-fold less than the α-tubulin gene. Still the transcriptional level of the EF-1α gene is not particularly lower than those of the vast majority of genes in Thalassiosira because the standard (α-tubulin) is a highly expressed gene. The expression patterns of the two genes were not significantly altered by the light and dark conditions (data not shown). However, Phaeodactylum EF-1α gene showed ≈5-fold higher expression than the α-tubulin gene (Table 1).
The drastic difference in expression detected between EF-1α and EFL genes (Table 1) may reflect the difference in cellular functions between the two proteins in Thalassiosira. The products of the highly expressed EFL gene may catalyze peptide chain elongation in Thalassiosira, as originally proposed in ref. 9. Neither the P/TKK motif nor the K indel directly overlapped with the putative residues for EF functions in the tertiary structure (Fig. S2), suggesting these motifs unlikely disrupt EF functions. However, the K indel position corresponds to a part of the GTP binding pocket in the EF-1α structure, so we carefully need to evaluate the influence of this motif on the property of GDP → GTP exchange in EFL in the future. To the contrary, EF-1α, the transcripts of which are present in much less abundance than those of the EFL gene, likely lost EF functions and operates the auxiliary functions (or a subset of these) originally possessed (1, 23, 24). In line with this possibility, it is intriguing that an EF-1α phylogeny showed a faster substitution rate of the Thalassiosira homologue than that of the Phaeodactylum homologues, which may function as the canonical EF (Fig. S1). The large rate difference between the two diatom homologues suggests that the Thalassiosira homologue receives fewer functional constraints than the Phaeodactylum counterpart and is congruent with the partial loss of original EF-1α functions proposed above.
We can provide additional evidence for the loss of EF functions in Thalassiosira EF-1α. Thalassiosira genome encodes no detectable EF-1β gene, the gene product of which is indispensable for GDP → GTP exchange of EF-1α during peptide elongation (1). This observation strongly suggests that (i) Thalassiosira EF-1α is not involved in peptide elongation and (ii) the putative auxiliary functions taken on by Thalassiosira EF-1α require no GTP hydrolysis. Phaeodactylum genome contains a canonical EF-1β gene (www.jgi.doe.gov), indicating the typical GDP → GTP exchange of EF-1α is catalyzed by EF-1β in this diatom cells. Unfortunately, it is unclear how EFL operates GDP → GTP exchange. No EF-1β gene has been identified in the complete genomes of Chlamydomonas or Ostreococcus, which also contain only EFL (www.jgi.doe.gov). EFL is likely a self-GTP recharging protein, or might interact with a subunit bearing no detectable sequence homology to known EF-1β. Precise EFL functions needs to be tested by using biochemical techniques in the future.
EF-1α and EFL genes have been believed to be mutually exclusive (9–12). To rationalize such EF-1α/EFL distribution, an unknown mechanism whereby the endogenous EF-1α gene is expelled by the exogenous EFL gene was hypothesized. However, the diatom data presented above clearly suggest that the two genes can coexist in a single organism, and we expect that additional dual-type eukaryotes distantly related to diatoms will be identified in the future. Likewise, secondary EFL gene loss, which is proposed so far for Phaeodactylum (this study) and a few lineages in Streptophyta (10), may be more common in eukaryotic evolution than we thought. If the above predictions are the case, no mechanism that promotes an EF-1α-to-EFL transition is necessary—after lateral EFL gene transfer, some lineages randomly retain either EF-1α or EFL, whereas other lineages keep both EFs through functional divergence of EF-1α (see above). Comparative biochemistry of EF-1α and EFL functions in distantly related dual-type lineages will be extremely intriguing, because the detailed functional divergence of the two EFs could be varied on a lineage-specific basis.
Materials and Methods
Strains.
Achnanthes kuwaitensis (NIES1349), Asterionella glacialis (NIES417), Chaetoceros didymus (NIES586), Cylindrotheca closterium (NIES1045), Ditylum brightwellii (NIES350), Skeletonema costatum (NIES17), and Thalassionema nitzschioides (NIES534) were purchased from the National Institute for Environmental Study. Detonula confervacea (CCMP353), Thalassiosira pseudonana (CCMP1335), and Phaeodactylum tricornutum (CCMP638) were purchased from the Provasoli-Guillard National Center for Culture of Marine Phytoplankton.
RNA Extraction and cDNA Synthesis.
Total RNA extraction was performed by using the RNeasy Plant Mini kit (QIAGEN) following the manufacturer's instruction. RT reactions were performed by using the Perfect Real-Time kit with random hexamer primers (TaKaRa), or the 3′ Full RACE Core kit with the poly(dT) primer (TaKaRa) following the manufacturer's instructions.
PCR, Cloning, and Sequencing.
Approximately 250-bp DNA fragments corresponding to the N termini of EF-1α and EF-1α-related proteins were amplified by the combination of one of three forward primers (5′-GGCCACGTGGAYTCNGGNAARTCNAC, 5′-GGCCACGTGGAYAGYGGNAARTCNAC, or 5′-GGCCACGTGGAYGCNGGNAARTCNAC) and a reverse primer (5′-ACGAAA T C T CTCTTRTGNCCNGGNGCRTC). The amplifications were consisted of 30 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min. PCR products were cloned into pGEM T-Easy vector (Promega). For each product, 9–15 clones were randomly sequenced on both strands. For subsequent 3′ RACE experiments, exact-matched forward primers were prepared based on the sequences of the initially amplified products. DNA fragments amplified from the 3′ RACE experiments were cloned and sequenced as described above.
Real-Time RT-PCR Assay.
Exact-matched primers for EF-1α, EFL, and α-tubulin for real-time PCR assay were designed based on the genome sequences of Thalassiosira and Phaeodactylum by the Joint Genome Institute (www.jgi.doe.gov). Standards for RT real-time PCR were generated as follows. The amplicon for each gene was amplified from the cDNA sample (synthesized with random hexamer primers) under the conditions of 94°C for 4 min followed by 30 cycles of 94°C for 30 sec, a gene-specific temperature (summarized in Table S2) for 30 sec, and 72°C for 30 sec. Target gene-fragments were cloned and verified by sequencing.
A mixture for real-time PCR contained SYBR Green I (TaKaRa), Premix Ex Taq (TaKaRa), a set of primers (final concentration of 0.3 μM each) (Table S2), and template solution—cDNA, the RNA sample (the negative control), or five differently diluted plasmid solutions, including 10 to 107 copies of the target gene fragments as the standards. We confirmed that a single target product was amplified by real-time PCR, based on melting curves (data not shown). In each assay, the target amplification from the RNA sample was out of the quantifiable range. Smart Cycler II (Cepheid) was used for all PCR described above.
Phylogenetic Analysis.
An EFL dataset was generated by adding the new sequences to the core alignment provided by M. Sakaguchi (University of Tsukuba, Tsukuba, Japan). The final EFL dataset with 318 unambiguously aligned amino acid positions was subjected to maximum-likelihood (ML) analyses, using IQPNNI software, Version 3.1 (25), and PhyML software, Version 2.4.4 (26). The amino acid substitutions in the dataset were modeled under the WAG model (27), incorporating among-site rate variation approximated by discrete gamma distributions with four categories (WAG+Γ model). ML bootstrap analyses (100 replicates) were conducted as described above.
The EFL dataset was also subjected to Bayesian analysis under the WAG+Γ model, using MrBayes software, Version 3.1.1 (28). One cold and three heated Markov chain Monte Carlo chains with default chain temperatures were run for 5 × 106 generations, sampling log-likelihoods and trees at 100-generation intervals. The first 1 × 104 generations (i.e., 100 trees) were discarded as “burn-in,” and posterior probabilities and branch lengths were obtained from the remaining trees.
Supplementary Material
Acknowledgments.
We thank G. Gile, P. J. Keeling (University of British Columbia, Vancouver, Canada), T. Amano (Kyoto University, Kyoto, Japan), K. Takishita (Japan Agency for Marine–Earth Science and Technology, Yokosuka, Japan), M. Sakaguchi (University of Tsukuba), and S. Mayama (Tokyo Gakugei University, Tokyo, Japan) for valuable discussions. We also thank J. B. Dacks (University of Cambridge, Cambridge, U.K.) for the critical reading of the manuscript. R.K. is a research fellow by the Japan Society for Promotion of Young Scientist (no. 1803336). This work was supported by the Japan Society for Promotion of Science Grant 18570214 (to Y.I.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank, European Molecular Biology Laboratory, and DNA Data Bank of Japan databases (accession nos. AB368767–AB368774).
This article contains supporting information online at www.pnas.org/cgi/content/full/0711084105/DCSupplemental.
References
- 1.Negrutskii BS, El'skaya AV. Eukaryotic translation elongation factor 1α: Structure, expression, functions, and possible role in aminoacyl-tRNA channeling. Prog Nucleic Acid Res Mol Biol. 1998;60:47–78. doi: 10.1016/s0079-6603(08)60889-2. [DOI] [PubMed] [Google Scholar]
- 2.Dreher TW, Uhlenbeck OC, Browning KS. Quantitative assessment of EF-1α. GTP binding to aminoacyl-tRNAs, aminoacyl-viral RNA, and tRNA shows close correspondence to the RNA binding properties of EF-Tu. J Biol Chem. 1999;274:666–672. doi: 10.1074/jbc.274.2.666. [DOI] [PubMed] [Google Scholar]
- 3.Kamaishi T, et al. Protein phylogeny of translation elongation factor EF-1α suggests microsporidians are extremely ancient eukaryotes. J Mol Evol. 1996;42:257–263. doi: 10.1007/BF02198852. [DOI] [PubMed] [Google Scholar]
- 4.Baldauf SL, Doolittle WF. Origin and evolution of the slime molds (Mycetozoa) Proc Natl Acad Sci USA. 1997;94:12007–12012. doi: 10.1073/pnas.94.22.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iwabe N, Kuma K, Hasegawa M, Osawa S, Miyata T. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci USA. 1989;86:9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inagaki Y, Doolittle WF, Baldauf SL, Roger AJ. Lateral transfer of an EF-1α gene: Origin and evolution of the large subunit of ATP sulfurylase in eubacteria. Curr Biol. 2002;12:772–776. doi: 10.1016/s0960-9822(02)00816-3. [DOI] [PubMed] [Google Scholar]
- 7.Inagaki Y, Susko E, Roger AJ. Recombination between elongation factor 1α genes from distantly related archaeal lineages. Proc Natl Acad Sci USA. 2006;103:4528–4533. doi: 10.1073/pnas.0600744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ke D, et al. Evidence for horizontal gene transfer in evolution of elongation factor Tu in enterococci. J Bacteriol. 2000;182:6913–6920. doi: 10.1128/jb.182.24.6913-6920.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keeling PJ, Inagaki Y. A class of eukaryotic GTPase with a punctate distribution suggesting multiple functional replacements of translation elongation factor 1α. Proc Natl Acad Sci USA. 2004;101:15380–15385. doi: 10.1073/pnas.0404505101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noble GP, Rogers MB, Keeling PJ. Complex distribution of EFL and EF-1α proteins in the green algal lineage. BMC Evol Biol. 2007;7:82. doi: 10.1186/1471-2148-7-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gile GH, Patron NJ, Keeling PJ. EFL GTPase in cryptomonads and the distribution of EFL and EF-1α in chromalveolates. Protist. 2006;157:435–444. doi: 10.1016/j.protis.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Ruiz-Trillo I, Lane CE, Archibald JM, Roger AJ. Insights into the evolutionary origin and genome architecture of the unicellular opisthokonts Capsaspora owczarzaki and Sphaeroforma arctica. J Eukaryot Microbiol. 2006;53:379–384. doi: 10.1111/j.1550-7408.2006.00118.x. [DOI] [PubMed] [Google Scholar]
- 13.Harper JT, Waanders E, Keeling PJ. On the monophyly of chromalveolates using a six-protein phylogeny of eukaryotes. Int J Syst Evol Microbiol. 2005;55:487–496. doi: 10.1099/ijs.0.63216-0. [DOI] [PubMed] [Google Scholar]
- 14.Kooistra WH, Medlin LK. Evolution of the diatoms (Bacillariophyta). IV. A reconstruction of their age from small subunit rRNA coding regions and the fossil record. Mol Phylogenet Evol. 1996;6:391–407. doi: 10.1006/mpev.1996.0088. [DOI] [PubMed] [Google Scholar]
- 15.Medlin LK, Kooistra WH, Gersonde R, Wellbrock U. Evolution of the diatoms (Bacillariophyta). II. Nuclear-encoded small-subunit rRNA sequence comparisons confirm a paraphyletic origin for the centric diatoms. Mol Biol Evol. 1996;13:67–75. doi: 10.1093/oxfordjournals.molbev.a025571. [DOI] [PubMed] [Google Scholar]
- 16.Ehara M, Inagaki Y, Watanabe KI, Ohama T. Phylogenetic analysis of diatom coxI genes and implications of a fluctuating GC content on mitochondrial genetic code evolution. Curr Genet. 2000;37:29–33. doi: 10.1007/s002940050004. [DOI] [PubMed] [Google Scholar]
- 17.Takishita K, Inagaki Y, Tsuchiya M, Sakaguchi M, Maruyama T. A close relationship between Cercozoa and Foraminifera supported by phylogenetic analyses based on combined amino acid sequences of three cytoskeletal proteins (actin, α-tubulin, and β-tubulin) Gene. 2005;362:153–160. doi: 10.1016/j.gene.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Burki F, et al. Phylogenomics reshuffles the eukaryotic supergroups. PLoS ONE. 2007;2:e790. doi: 10.1371/journal.pone.0000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hackett JD, et al. Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of rhizaria with chromalveolates. Mol Biol Evol. 2007;24:1702–1713. doi: 10.1093/molbev/msm089. [DOI] [PubMed] [Google Scholar]
- 20.Adl SM, et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- 21.Lee MJ, Ellis R, Lee JJ. A comparative study of photoadaptation in four diatoms isolated as endosymbionts from larger foraminifera. Mar Biol. 1982;68:193–197. [Google Scholar]
- 22.Mayama S, Nagumo T, Kuriyama A. Isolation and identification of endosymbiotic diatoms from planktonic and benthic species of foraminifera. Diatom. 2000;16:3–10. [Google Scholar]
- 23.Liu G, Edmonds BT, Condeelis J. pH, EF-1α and the cytoskeleton. Trends Cell Biol. 1996;6:168–171. doi: 10.1016/0962-8924(96)20013-3. [DOI] [PubMed] [Google Scholar]
- 24.Gonen H, et al. Protein synthesis elongation factor EF-1α is essential for ubiquitin-dependent degradation of certain N α-acetylated proteins and may be substituted for by the bacterial elongation factor EF-Tu. Proc Natl Acad Sci USA. 1994;91:7648–7652. doi: 10.1073/pnas.91.16.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vinh le S, Von Haeseler A. IQPNNI: Moving fast through tree space and stopping in time. Mol Biol Evol. 2004;21:1565–1571. doi: 10.1093/molbev/msh176. [DOI] [PubMed] [Google Scholar]
- 26.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 27.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 28.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.