Abstract
The recent identification of the genes responsible for several human genetic diseases affecting bone homeostasis and the characterization of mouse models for these diseases indicated that canonical Wnt signaling plays a critical role in the control of bone mass. Here, we report that the osteoblast-specific transcription factor Osterix (Osx), which is required for osteoblast differentiation, inhibits Wnt pathway activity. First, in calvarial cells of embryonic day (E)18.5 Osx-null embryos, expression of the Wnt antagonist Dkk1 was abolished, and that of Wnt target genes c-Myc and cyclin D1 was increased. Moreover, our studies demonstrated that Osx bound to and activated the Dkk1 promoter. In addition, Osx inhibited β-catenin-induced Topflash reporter activity and β-catenin-induced secondary axis formation in Xenopus embryos. Importantly, in calvaria of E18.5 Osx-null embryos harboring the TOPGAL reporter transgene, β-galactosidase activity was increased, suggesting that Osx inhibited the Wnt pathway in osteoblasts in vivo. Our data further showed that Osx disrupted binding of Tcf to DNA, providing a likely mechanism for the inhibition by Osx of β-catenin transcriptional activity. We also showed that Osx decreased osteoblast proliferation. Indeed, E18.5 Osx-null calvaria showed greater BrdU incorporation than wild-type calvaria and that Osx overexpression in C2C12 mesenchymal cells inhibited cell growth. Because Wnt signaling has a major role in stimulating osteoblast proliferation, we speculate that Osx-mediated inhibition of osteoblast proliferation is a consequence of the Osx-mediated control of Wnt/β-catenin activity. Our results add a layer of control to Wnt/β-catenin signaling in bone.
Bone formation takes place through two distinct processes. Most of the skeleton is formed by endochondral ossification involving a cartilage model. A small number of skeletal elements, mainly craniofacial bones, are formed by intramembranous ossification, by which bones form directly from condensations of mesenchymal cells without a cartilage intermediate. The bone-forming osteoblasts and the cartilage-forming chondrocytes are derived from a common mesenchymal progenitor (1). Osteoblast differentiation occurs through a multistep molecular pathway regulated by different transcription factors and signaling proteins. Indian hedgehog (Ihh), a member of the Hedgehog family, which is required for endochondral but not for intramembranous bone formation (2), is needed for the establishment of the osteogenic portion of the perichondrium/periosteum and for the initial activation of the gene for Runx2, which is required for the differentiation of mesenchymal cells into preosteoblasts. The transcription factor Runx2 is needed for the formation of both endochondral and membranous skeletal elements. Indeed, in Runx2-null mutants, no endochondral and no membranous bones form, and osteoblast differentiation is arrested at an early step of bone development (3). Heterozygous mutations in Runx2 are the cause of the human genetic disease cleidocranial dysplasia (4). Runx2 is also needed for the expression of another transcription factor Osterix (Osx), which is required for the differentiation of preosteoblasts into functional osteoblasts. Osx is specifically expressed in all osteoblasts and at low levels in prehypertrophic chondrocytes. In Osx-null embryos, a full cartilage skeleton is formed, but the embryos completely lack bone formation. In these embryos, osteoblast differentiation markers, such as bone sialoprotein, osteonectin, and osteocalcin, are not expressed (5). The DNA-binding domain of Osx is located at its C terminus and consists of three C2H2-type zinc fingers that share a high degree of identity with a similar motif in Sp1, Sp3, and Sp4. Osx binds to specific GC-rich sequences and contains a proline-rich transcription activation domain.
The canonical Wnt pathway is controlled both outside and inside the cell. Wnt polypeptides bind to frizzled receptors and LRP5/6 coreceptors (6). Several antagonists, including Dkks, sclerostin, and soluble frizzled receptors, inhibit Wnt signaling (7). The essential event in Wnt signaling is the accumulation and nuclear translocation of β-catenin, which then interacts with members of the Lef/Tcf family of transcription factors to activate target genes (7). Distinct Wnts control early events during skeletal development such as limb patterning and joint formation (8).
Wnt/β-catenin signaling, in addition to the transcription factors Runx2 and Osx, is essential to osteoblast differentiation during embryonic development. Conditional inactivation of β-catenin in either skeletal progenitor cells or at a later stage of osteoblast development in mouse embryos blocks osteoblast differentiation (9–12). When β-catenin is inactivated in early skeletal progenitors, low levels of Runx2, but not Osx, expression are detected in these cells (11). At a later step, however, the inactivation of β-catenin does not prevent the expression of Osx, indicating that Wnt signaling is not needed for Osx expression at this step (12).
Several lines of evidence indicate that canonical Wnt signaling is also required for normal osteoblast proliferation. First, if β-catenin is stabilized in osteoblasts during mouse embryonic development, a marked increase in osteoblast proliferation occurs (12). Moreover, Lrp5-null mice, which phenocopy the osteoporosis-pseudoglioma syndrome in humans (13), develop a phenotype with low bone mass because of decreased osteoblast proliferation (14). In contrast, gain-of-function mutants of Lrp5 cause high bone mass syndrome in patients (15) and in mice (16). In addition, the Wnt signaling antagonist Dkk1 prevents the activation of Wnt signaling by binding to LRP5/6. The bone formation and bone mass of heterozygous Dkk1 mutant mice increase with an increased number of osteoblasts (17). In contrast, the overexpression of Dkk1 in osteoblasts leads to severe osteopenia with decreased osteoblast numbers (18). Thus, Wnt/β-catenin signaling stimulates osteoblast proliferation.
In other genetic experiments using mice in which a stabilized β-catenin was expressed in mature osteoblasts, the expression of osteoprotegerin, a decoy receptor for RANK ligand, was increased. This resulted in a decrease in osteoclast differentiation and function, an inhibition of bone degradation, and increased bone mass (19, 20). In contrast, in mice in which the β-catenin gene was ablated in mature osteoblasts, an osteopenic phenotype and lower level of osteoprotegerin were observed (19, 20). These experiments indicate that activation of β-catenin in osteoblasts noncell autonomously inhibits osteoclast differentiation and function.
The experiments reported in this article strongly support the hypothesis that, in addition to its essential role in osteoblast differentiation, the osteoblast-specific transcription factor Osx negatively regulates Wnt/β-catenin signaling and osteoblast proliferation.
Results
Effect of Osx on Osteoblast Proliferation.
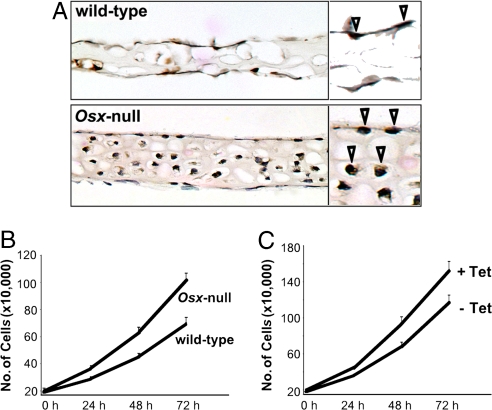
To further characterize Osx-null embryos, we examined whether osteoblast proliferation was affected in these mutants in vivo. To this end, we measured the number of cells actively synthesizing DNA by 5-bromo-2′-deoxy-uridine (BrdU) incorporation and compared the percentage of BrdU-positive cells in the calvaria of embryonic day (E)18.5 Osx-null and Osx wild-type embryos. Fig. 1A shows that BrdU incorporation was greater in calvaria of Osx-null E18.5 embryos than in wild-type calvaria. Primary calvarial cells from Osx-null E18.5 embryos also grew faster than wild-type cells (Fig. 1B). Therefore, these experiments suggest that Osx inhibits osteoblast growth. To further test this possibility, we generated stable C2C12 mesenchymal cells in which the overexpression of Osx could be induced by using the Tet-off system. Although Osx was not expressed in the presence of Tet, Osx was highly expressed in the absence of Tet in the stable cell lines (data not shown). This induction of Osx expression in turn inhibited cell growth (Fig. 1C). Overall, these observations indicate that Osx has the ability to control cell proliferation.
Fig. 1.
Effect of Osx on cell proliferation. (A) BrdU incorporation in sections of calvaria from E18.5 wild-type and Osx-null embryos. Pregnant females were injected i.p. with BrdU 4 h before being killed. Sections were obtained from four wild-type and four Osx-null embryos, and BrdU-positive cells were counted in 10 different fields. The percentages of BrdU positive cells were 9.2 ± 1.4 for wild-type calvaria and 45.8 ± 2.8 for Osx-null calvaria. Arrowhead in the insert indicate BrdU-positive cells. (B) Cell growth of E18.5 wild-type and Osx-null primary calvaria cells. (C) A stably transfected C2C12 cell line was generated in which the expression of Osx could be induced by using the Tet-off system in the absence of tetracycline. Comparison of cell growth in the presence (+) or absence (−) of tetracycline.
Wnt Pathway Signaling Is Down-Regulated by Osx.
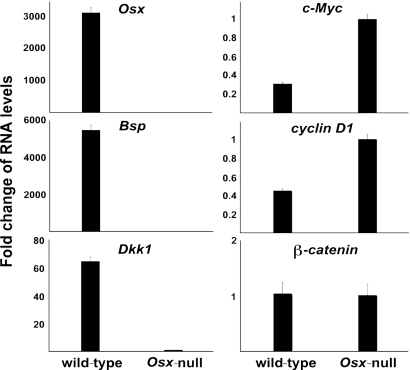
To explore possible mechanisms by which Osx controls osteoblast proliferation, we used microarrays to compare the RNA expression profiles of wild-type and Osx-null calvarial cells from E18.5 embryos. These experiments revealed the broad repertoire of genes that constitute the genetic program of osteoblasts controlled by Osx and strongly support the view that Osx is a major effector of the osteoblast program (C.Z. and B.d.C., unpublished data). Fig. 2 compares the levels of expression of a discrete number of calvarial RNAs in E18.5 wild-type and Osx-null embryos as measured by quantitative RT-PCR. As expected, the expression of the osteoblast marker bone sialoprotein was abolished in Osx-null calvarial cells. Among the differentially expressed genes, the expression of the canonical Wnt signaling inhibitor Dkk1 was abolished, but β-catenin expression did not change. In contrast, expression of several Wnt target genes, such as c-Myc and cyclin D1, was greater in Osx-null calvarial cells than in wild-type cells. Expression of Cdk4 was also increased in Osx-null calvarial cells, whereas expression of cyclin D2 was unchanged (data not shown). In E18.5 Osx-null calvarial cells, expressions of both OPG and RANKL were down-regulated as measured by quantitative RT-PCR, but the ratio of OPG/RANKL increased. Expression of the osteoclast marker TRAP was also down-regulated [supporting information (SI) Fig. S1A]. Although in E18.5 Osx-null limbs, there were TRAP positive cells, the number of these cells was less than in E18.5 wild-type embryos (Fig. S1 B and C). In long bones of Osx-null embryos, cells from the periosteum invade the zone of hypertrophy chondrocyte as a wedge-shaped expansion of the periosteum in which osteoblast precursors are arrested in their differentiation (5). Given the major difference between the skeletal element of Osx-null and wild-type E18.5 embryos, a precise comparison of the number of osteoclasts between mutant and wild-type is difficult to measure.
Fig. 2.
Fold change in RNA levels of specific calvaria RNAs from E18.5 Osx wild-type and Osx-null embryos. RNA levels were measured by real-time RT-PCR. The level of each type of RNA from Osx-null calvaria was normalized to a value of 1.
Osx Stimulates Dkk1 Promoter Activity.
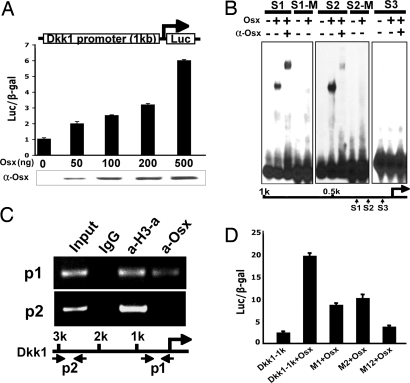
Because both microarray and real-time RT-PCR results suggested that Osx is required for Dkk1 expression in osteoblasts, we asked whether Osx stimulates Dkk1 promoter activity. In transfection experiments of HEK293 cells, Osx activated a 1-kb Dkk1 promoter luciferase reporter in a dose-dependent manner (Fig. 3A), suggesting that Osx might transcriptionally activate the Dkk1 gene in osteoblasts. DNA sequence analysis revealed three potential Osx binding sites in a 1-kb Dkk1 promoter. In EMSA purified recombinant, Osx bound to sites of S1 and S2 (but not to S3) and anti-Osx antibody supershifted Osx-DNA complexes. In addition, S1 mutation S1-M and S2 mutation S2-M inhibited Osx binding, indicating that Osx specifically binds to sites of S1 and S2 (Fig. 3B). In primary calvarial cells from new-born wild-type mice, ChIP analysis showed that Osx was associated with the chromatin of a segment of the Dkk1 promoter covering S1, S2, and S3 sites, whereas no Osx was associated with the chromatin of a more 5′ segment of the Dkk1 gene (Fig. 3C). Mutation in either S1 or S2 inhibited Osx activation of the 1-kb Dkk1 promoter 50–60%, whereas mutations in both sites almost abolished activation of the promoter (Fig. 3D).
Fig. 3.
Osx activates a Dkk1 promoter. (A) HEK293 cells were transfected with a 1-kb Dkk1 promoter luciferase reporter without or with an increasing amount of pEX-Osx as indicated. Cell extracts were subjected to Western blot analysis to monitor the expression of transfected pEX-Osx. (B) Purified recombinant Osx binds to two specific sites in the Dkk1 promoter in EMSA. Three potential Osx binding sites in the 1-kb Dkk1 promoter, S1, S2, and S3, were used as probes. S1-M is S1 site mutation and S2-M is S2 site mutation. (C) Osx associates with the chromatin of a segment of the Dkk1 promoter in ChIP assay. Calvarial cells were isolated and cultured from wild-type new born mice. p1 primer set corresponds to a segment of the Dkk1 promoter covering S1, S2, and S3 sites within the proximal 500 bp. As a negative control, p2 primer set covers a distal region of the Dkk1 promoter, which does not contain an Osx binding site. Anti-acetylated histone H3 antibody (a-H3-a) was used as a positive control. (D) Mutations in sites S1 and S2 inhibit activation of the Dkk1 promoter by Osx. Mutations in the 1-kb Dkk1 reporter were constructed. Mutant M1 contains the S1 site mutation, mutant M2 the S2 site mutation, and double mutant M12 (both S1 and S2 mutations) in the 1-kb Dkk1 promoter. HEK293 cells were transfected with wild-type and mutant Dkk1 promoter reporters without or with pEX-Osx, as indicated.
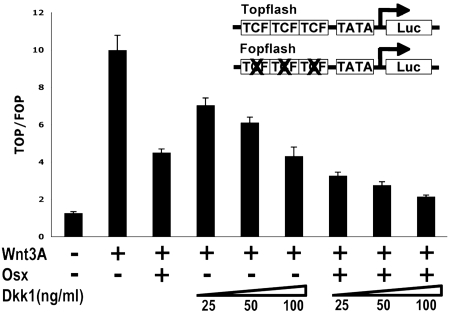
Topflash is a reporter construct activated by Wnt signaling and, hence, by Wnt3A-conditioned medium. As shown in Fig. 4, the addition of recombinant Dkk1 inhibited the activity of Wnt3A-induced Topflash reporter in a dose-dependent manner. The transfection of Osx also inhibited Wnt3A-induced Topflash reporter activity. The transfection of Osx together with the addition of recombinant Dkk1 further inhibited the Wnt3A-induced Topflash reporter, suggesting that Osx and Dkk1 have an additive effect in the inhibition of Topflash reporter activity. These results raised the possibility that Osx might target additional components of the Wnt pathway.
Fig. 4.
Cooperation between Osx and Dkk1 in the inhibition of Wnt3A-induced Topflash activity. Topflash and Fopflash reporters were transfected in HEK293 cells with pEX-Osx as indicated. Wnt3A-conditioned medium was used to activate the Topflash reporter. Recombinant Dkk1 protein was added to the medium at the different concentrations as indicated. Results were expressed as the ratio of Topflash over Fopflash activity.
Effect of Osx on Wnt Signaling Activity.
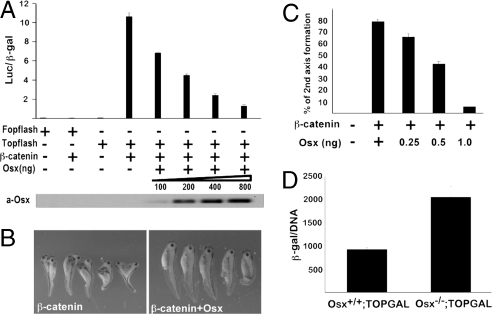
To begin to identify other possible Wnt pathway components targeted by Osx, we took advantage of the established model of β-catenin-induced Topflash activation. Osx strongly inhibited β-catenin-induced Topflash reporter expression in a dose-dependent manner (Fig. 5A). This was further confirmed by in vivo results demonstrating that Osx inhibited β-catenin-induced secondary axis formation in Xenopus embryos in a dose-dependent manner (Fig. 5 B and C). To examine whether Wnt signaling in E18.5 mouse skeletal elements was increased in vivo in the absence of Osx, we used the Wnt indicator TOPGAL transgenic mice. The TOPGAL transgene consists of a β-galactosidase gene driven by a Tcf/β-catenin-responsive promoter, which enables canonical Wnt activity to be measured by β-galactosidase activity in extracts of intact embryonic tissues. In E18.5 Osx−/− embryos harboring the TOPGAL transgene, the calvarial β-galactosidase activity per unit of DNA was increased approximately twofold compared with that in Osx+/+;TOPGAL embryos (Fig. 5D). These experiments provide evidence that Osx inhibits the transcriptional activity of β-catenin. Furthermore, the increase in TOPGAL activity in the calvaria of Osx-null embryos implies that the inhibition of Wnt/β-catenin signaling occurs in osteoblasts in vivo. Overall, our experiments suggest that Osx has the ability to activate expression of the canonical Wnt signaling antagonist Dkk1 to directly inhibit the transcriptional activity of β-catenin.
Fig. 5.
Inhibition of β-catenin transcription activity by Osx. (A) Osx inhibits β-catenin-induced Wnt reporter activity. HEK293 cells were transfected with the Topflash or Fopflash reporter along with Myc-β-catenin, a plasmid expressing a stabilized β-catenin, and increasing amounts of pEX-Osx DNA. (B) Osx inhibits β-catenin-induced secondary axis formation in Xenopus embryos. Embryos were microinjected with RNA for stabilized β-catenin (100 pg) and increasing amounts of Osx RNA at the four-cell cleavage stage into the equatorial region of a single vegetal-ventral blastomere. Microinjection of β-catenin RNA in the ventral side of four-cell Xenopus embryos induces the formation of a secondary body axis. The phenotypes were evaluated by using a binocular dissecting microscope at the tadpole stage. (C) Dose-dependent inhibition of β-catenin-induced secondary axis formation in Xenopus embryos by Osx. (D) β-galactosidase activity in calvarial extracts of E18.5 Osx+/+;TOPGAL and Osx−/−;TOPGAL embryos. Results were expressed as the ratio of β- galactosidase over DNA.
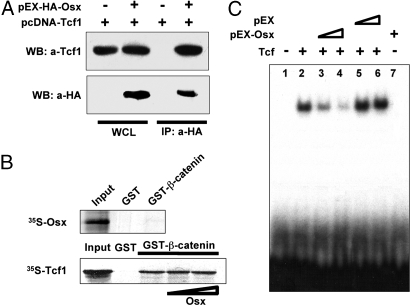
Because our results showed that Osx inhibited β-catenin-induced Topflash activity, we examined possible interactions of Osx with nuclear components of the canonical Wnt pathway. We first tested whether Osx and Tcf could associate with each other in the same complex. Among TCF family members, both Tcf1 and Tcf4 are expressed in osteoblasts. Tcf1 is detectable in osteoblasts from E14.5, whereas Tcf4 is detectable after E16.5. In cotransfections of HEK293 cells, Osx and Tcf1 were shown to coimmunoprecipitate (Fig. 6A). Osx was also shown to coimmunoprecipitate with Tcf4 (data not shown). In GST pull-down experiments, GST-β-catenin did not pull-down 35S-labeled Osx, but, as expected, it did pull down 35S-Tcf1. Bacculovirus-expressed Osx failed to disrupt the pull-down of 35S-Tcf1 by GST-β-catenin (Fig. 6B).
Fig. 6.
Interaction between Osx and Tcf1. (A) Coimmunoprecipitation (CoIP) of Osx and Tcf1 in transfected HEK293 cells. Here, 1 μg of pEX-HA-Osx and pCDNA-Tcf1 were cotransfected or transfected alone into HEK293 cells. Whole-cell lysates (WCL) were immunoprecipitated with anti-HA (10 μl) and the precipitate was immunoblotted with anti-Tcf1. (B) No disruption of β-catenin interaction with Tcf1 by Osx. (Upper) GST-β-catenin was used to pull down 35S labeled Osx. Ten percent of the synthesized Osx was used as an input, and GST was used as a control. (Lower) GST-β-catenin was used to pull down 35S-labeled Tcf1. Baculovirus-expressed Osx was used as Osx protein source. (C) Disruption of Tcf1 binding to DNA by Osx. Osx and Tcf1 proteins were synthesized by TNT IVTT systems. Tcf1 bound to Tcf 1 binding probe in EMSA. TNT lysates containing pEX-Osx and control TNT lysates with pEX were added to the DNA binding reaction.
Tcfs bind to the promoter region of Wnt target genes to regulate their expression. Because Osx did not disrupt β-catenin/Tcf complex formation, we assessed whether Osx might disrupt Tcf binding to DNA. Based on promoter sequence analysis, there is no Osx binding site overlapping the consensus Tcf binding sequences; however, it is possible that, by interacting with Tcf, Osx disrupts Tcf binding to DNA. In EMSA, Tcf1 bound to a 32P-labeled double-strand oligo containing a consensus Tcf binding site, whereas Osx generated by in vitro transcription and translation (IVTT) did not bind to this DNA. When increasing amounts of Osx were added, Tcf1 binding to DNA was disrupted in a dose-dependent manner, whereas the control IVTT product failed to affect Tcf1 binding to DNA (Fig. 6C).
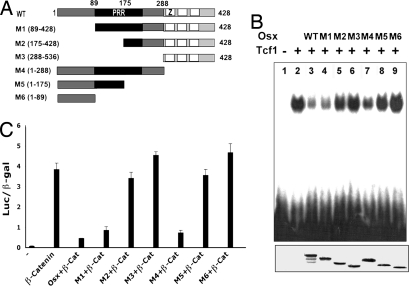
To determine which domain of Osx was responsible for the disruption of Tcf binding to DNA, a series of deletions was generated (Fig. 7A). Osx contains a proline-rich region (PRR) transactivation domain toward the N-terminal part of the protein and a three-zinc-finger DNA-binding motif in the C-terminal region. As shown in Fig. 7B, mutants containing PRR, such as M1 and M4, inhibited Tcf binding to DNA, whereas mutants lacking part or all of the PRR, such as M2, M3, M5, and M6, failed to disrupt Tcf binding to DNA. These results suggest that the PRR domain is responsible for the inhibition of Tcf binding to DNA. Next, we evaluated whether the PRR region was required for the Osx inhibitory effect on Wnt signaling activity. To address this question, Osx mutants were cotransfected with β-catenin into HEK293 cells along with the Topflash reporter. Fig. 7C shows that the Osx mutants with the PRR region, M1 and M4, were able to inhibit β-catenin-induced Topflash reporter, whereas the Osx mutants without a complete PRR region, M2, M3, M5 and M6, were not. Therefore, we conclude that the PRR region mediates both the Osx inhibition of β-catenin-induced Topflash reporter activation and the disruption of Tcf binding to its target DNA.
Fig. 7.
PRR region of Osx is needed for disruption of Tcf1 binding to DNA and for inhibition of β-catenin transcriptional activity. (A) Schematic representation of the Osx deletion mutants. PRR, proline-rich region; Z, zinc-finger domain. (B) Osx and Tcf1 proteins were synthesized by TNT IVTT systems. M1 and M4 are mutants with full PRR region. M2, M3, M5, and M6 are mutants without full PRR region. (Lower) [35S]methionine-labeled Osx wild-type and mutants synthesized by IVTT system were detected by autoradiograph. (C) Osx PRR domain is required for Osx inhibitory effect on Wnt pathway. Vectors expressing Osx wild-type and mutants were transfected in HEK293 cells with or without vectors expressing activated β-catenin (β-cat) with the Topflash reporter.
Discussion
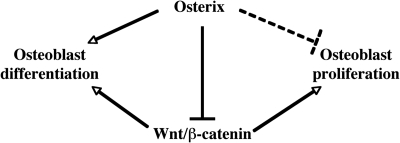
Here, we have presented evidence for two properties of the osteoblast-specific transcription factor Osx: that it inhibits Wnt signaling and decreases cell proliferation (Fig. 8). In Osx-null calvarial cells, the expression of the gene for the Wnt antagonist Dkk1 was abolished, suggesting that Osx positively controls the expression of Dkk1. This is also supported by the Osx-mediated activation of a 1-kb Dkk1 promoter fragment, the existence of two functional Osx binding sites in this promoter and the presence of Osx in the chromatin of this promoter in primary osteoblasts. In addition, both DNA transfection experiments and the inhibition of β-catenin-induced secondary axis formation in Xenopus embryos strongly suggested that Osx inhibits the transcriptional activity of β-catenin. In the transfection experiments, the proline-rich transcription activation domain of Osx was required for the inhibitory activity. Importantly, in vivo experiments showing that the Wnt-dependent TOPGAL promoter was more active in E18.5 Osx-null calvaria than in wild-type embryonic calvaria strongly suggested that Osx inhibits Wnt signaling in osteoblasts in vivo.
Fig. 8.
Proposed model of coordinated regulation of osteoblast differentiation and proliferation by Osx and Wnt/β-catenin signaling. Wnt/β-catenin signaling has an essential role in osteoblast differentiation during embryonic development and has a major role in stimulating osteoblast proliferation both during embryonic development and postnatally. Osx is an osteoblast-specific transcription factor, required for osteoblast differentiation. The inhibition of Wnt/β-catenin signaling activity by Osx also constitutes a possible mechanism for the inhibition by Osx of osteoblast proliferation. Not shown is the inhibitory role of Wnt/β-catenin in osteoclast differentiation and function.
In further transfection experiments, Osx and Tcf1, a DNA binding partner of β-catenin, were coimmunoprecipitated, which suggested that they are part of the same complex. In addition, the disruption of Tcf1 binding to DNA by Osx provided a likely mechanism for the inhibition of β-catenin transcriptional activity by Osx. Indeed, the same Osx mutants that failed to inhibit the transcriptional activity of β-catenin also lacked the ability to disrupt Tcf1 binding to DNA. Thus, our findings indicate that Osx negatively controls the activity of β-catenin in two ways: first, by being needed for the expression of a major Wnt antagonist, and second, by inhibiting the transcriptional activity of β-catenin/Tcf. Note that our results do not rule out the possibility that Osx might also target other components of Wnt signaling as part of its inhibitory effect.
The evidence for the ability of Osx to inhibit cell proliferation includes the markedly enhanced proliferation of E18.5 Osx-null calvarial cells compared to wild-type embryonic calvarial cells and the decrease in C2C12 cell proliferation after the induction of Osx expression. Other studies (12–18) provide strong genetic evidence that Wnt signaling positively regulates osteoblast proliferation. Therefore, we speculate that the Osx-mediated inhibition of osteoblast proliferation is a consequence of the inhibition of Wnt signaling by Osx. In C2C12 mesenchymal cells treated with amounts of Dkk1, which maximally inhibited cell proliferation, induction of Osx caused a further albeit modest reduction of cell growth (Fig. S2). Although even in the presence of saturating amounts of Dkk1 a residual activity of Wnt signaling may persist that could be further inhibited by disruption of Tcf binding to target genes by Osx, one cannot exclude that Osx might also inhibit other proproliferative pathways.
Our observation that Dkk1 expression is absent in Osx-null calvaria of E18.5 embryos parallels the finding reported in ref. 12 that Dkk1 expression was absent in tibia of E18.5 embryos in which β-catenin was inactivated by using an Osx-Cre transgene. This suggests that both Osx and β-catenin are required for Dkk1 expression in osteoblasts and that the Dkk1 gene might be a common target of these transcription factors in these cells. Furthermore, Osx is required for Dkk1 expression, which should further reinforce the feedback control effect that Dkk1 exerts on Wnt/β-catenin signaling.
Reports indicate that Wnt/β-catenin signaling can control bone mass by several mechanisms (9–12, 20). Indeed it has essential functions in (i) in osteoblast differentiation, (ii) proper osteoblast proliferation, and (iii) inhibiting bone resorption noncell autonomously by controlling osteoclast differentiation (9–12, 20). Mutations in LRP5, which result in either a high or low bone mass phenotype affect mainly osteoblast proliferation but do not produce an inhibition of osteoclast differentiation (14, 16). In contrast, either expression of a stabilized β-catenin or inactivation of β-catenin, specifically in mature osteoblasts in mice, which also cause high or low bone mass respectively, affect mainly osteoclast differentiation and function by controlling the expression in osteoblasts of osteoprotegrin, a decoy receptor for RANK ligand, without causing significant changes in osteoblast proliferation (19, 20). In the skeletal elements of E18.5 Osx-null embryos, the number of TRAP-positive cells appear to be reduced compared to wild-type embryos. This observation is supported by the increased OPG/RANKL ratio and the decreased TRAP mRNA in calvarial cell of E18.5 Osx-null embryos. Thus, it is possible that the inhibition of Wnt signaling by Osx also reduces osteoclast differentiation and function. We speculate that the inhibition of Wnt/β-catenin signaling by Osx, which itself has an essential role in osteoblast differentiation, ensures an optimal bone formation rate.
The extensive studies by many laboratories to understand the control of the Wnt signaling pathway in osteoblasts stems from the realization that this pathway has an essential role in bone mass determination in the adult skeleton. There is also an expectation that efforts to pharmacologically target this pathway should yield promising agents to treat bone diseases, such as osteoporosis. Our results showing that Osx inhibits Wnt/β-catenin signaling add an important new layer of control to the complex regulation of the Wnt pathway in osteoblasts (Fig. 8).
Materials and Methods
Plasmids, Protein Purification, Cell Culture, and Transient Transfections.
Osx deletion mutants were generated by PCR and subcloned into the pEX vector for transfection studies or into the pCS2+ vector for Xenopus experiments. Dkk1 reporter mutants were made with the QuikChange site-directed mutagenesis kit (Stratagene). All constructs were verified by DNA sequencing. GST-β-catenin was expressed and purified by glutathione-agarose affinity chromatography, and the in vitro protein interaction assay was performed as described in ref. 21. Osx cDNA was subcloned into the pBac vector, and recombinant baculovirus-expressed Osx was purified by nickel affinity chromatography (21). [35S]methionine-labeled protein was synthesized in a TNT T7 coupled in vitro transcription-translation system, using rabbit reticulocyte lysate (Promega). HEK293 cells were cultured in DMEM supplemented with 10% FBS and transfected by FuGENE 6 (Roche). For Topflash reporter assays, 250 ng of reporters and 50 ng of pSV2-βgal were cotransfected with the indicated expression plasmids. Luciferase activities were normalized for transfection efficiency to β-galactosidase activity. Conditioned media were from Wnt-3A-expressing mouse and parental L cells (American Type Culture Collection) (22).
RNA Isolation and Quantitative RT-PCR.
Total RNA was isolated with TRIzol reagent (Invitrogen). RNA was subjected to quantitative RT-PCR, using the TaqMan One-Step RT-PCR Master Mix reagent (Applied Biosystems). Relative transcript levels were measured by real-time PCR in a 50-μl reaction volume on 96-well plates, using an ABI PRISM 7000 sequence detection system (Applied Biosystems). Transcript levels were normalized to glyceraldehyde-3-phosphate dehydrogenase levels, using primers from Applied Biosystems.
Histological Analysis and Xenopus laevis Secondary-Axis Assays.
To examine cell proliferation, pregnant female mice were injected i.p. with 100 μl of 100 μM BrdU and killed 4 h later. Calvaria from E18.5 embryos were isolated, fixed, and processed. Sections of 5-μm were prepared and stained with the BrdU staining kit (Zymed). TRAP staining was performed in limb sections, using a leucocyte acid phosphatase kit (Sigma). X. laevis embryo microinjections were performed as described in ref. 22. Briefly, pCS2+-Osx and pCS2+-β-catenin were linearized and transcribed in vitro into capped mRNA, using the SP6 mMACHINE kit (Ambion). Xenopus females were injected with 800 units of human CG 10–16 h before manual squeezing to isolate eggs. The eggs were fertilized in vitro. Embryos were microinjected at the four-cell cleavage stage into the equatorial region of a single vegetal-ventral blastomere and placed in a solution of 5% Ficoll in 1× Marc's Modified Ringers (MMR) for 60–90 min before transfer to 0.1× MMR. mRNAs were injected at a volume of 10 nl per blastomere. Embryonic phenotypes were evaluated by using a standard binocular dissecting microscope (Nikon; SMZ-U) and captured as digital images (Nikon; CoolPix 995) at the tadpole stage.
Chromatin Immunoprecipitation (ChIP) Assay.
ChIP assays were performed as described in ref. 23 with some modifications. Briefly, Calvarial cells were isolated from wild-type newborn mice and were cultured in DMEM supplemented with 10% FBS. Formaldehyde was used to cross-link the cells for 10 min, and cross-linking was quenched with glycine. Cells were harvested and rinsed with PBS, and cell pellets were resuspended in 1 ml of lysis buffer. After sonication, 100 μl of sheared chromatin was diluted to 1 ml with IP dilution buffer for each immunoprecipitation. The chromatin solution was precleared with 60 μl of protein G-agarose beads at 4°C for 1 h. The precleared chromatin was collected and incubated at 4°C overnight with 5 μg of anti-Osx antibody, anti-acetylated histone H3 antibody (Upstate Biotechnology) or Rabbit IgG (Sigma) as a negative control. The immune complexes were precipitated with 60 μl of protein G-agarose beads at 4°C for 1 h. After washes, the antibody-protein-DNA immunocomplexes were eluted twice with 100 μl of elution buffer. Formaldehyde cross-linking was reversed by heating at 65°C overnight with the addition of 5 M NaCl. All of the samples were digested with RNase A and proteinase K. The DNA was purified by using spin columns, and analyzed by PCR. The p1 primer sequences for the Dkk1 promoter were 5′-CTAGTGCTCTAGTGACCCACACTC-3′ and 5′-TGGACTGCGGAACCTCAACTTC-3′. The p2 sequences were 5′-CACTCCATTGCCTGGCTGCATG-3′ and 5′-CATGGTATCAGGTCACAGAGGGAG-3′.
Supporting Information.
Details of immunoprecipitation, western blot analysis, the in vitro protein interaction assay, and EMSA may be found in SI Text.
Supplementary Material
Acknowledgments.
We thank Hui Dai, Lingna Zhang, and Dr. Francoise Coustry for technical assistance and Dr. Hans Clevers (Hubrecht Laboratory, Utrecht, The Netherlands) for providing the plasmids pcDNA-Tcf1 and pcDNA-Tcf4. This work was supported by National Institutes of Health Grant R01AR 49072 (to B.d.C.), an Arthritis Foundation Postdoctoral Fellowship (to C.Z.), and National Institutes of Health Grant CA16672 (for DNA sequence analysis).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710831105/DCSupplemental.
References
- 1.Akiyama H, et al. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc Natl Acad Sci USA. 2005;102:14665–14670. doi: 10.1073/pnas.0504750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komori T, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 4.Mundlos S, et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 5.Nakashima K, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 6.He X, Semenov M, Tamai K, Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: Arrows point the way. Development (Cambridge, UK) 2004;131:1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- 7.Krishnan V, Bryant HU, Macdougald OA. Regulation of bone mass by Wnt signaling. J Clin Invest. 2006;116:1202–1209. doi: 10.1172/JCI28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartmann C, Tabin CJ. Wnt-14 plays a pivotal role in inducing synovial joint formation in the developing appendicular skeleton. Cell. 2001;104:341–351. doi: 10.1016/s0092-8674(01)00222-7. [DOI] [PubMed] [Google Scholar]
- 9.Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 10.Hill TP, Spater D, Taketo MM, Birchmeier W, Hartmann C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell. 2005;8:727–738. doi: 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Hu H, et al. Sequential roles of Hedgehog and Wnt signaling in osteoblast development. Development (Cambridge, UK) 2005;132:49–60. doi: 10.1242/dev.01564. [DOI] [PubMed] [Google Scholar]
- 12.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development (Cambridge, UK) 2006;133:3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 13.Gong Y, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 14.Kato M, et al. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol. 2002;157:303–314. doi: 10.1083/jcb.200201089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Little RD, et al. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am J Hum Genet. 2002;70:11–19. doi: 10.1086/338450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Babij P, et al. High bone mass in mice expressing a mutant LRP5 gene. J Bone Miner Res. 2003;18:960–974. doi: 10.1359/jbmr.2003.18.6.960. [DOI] [PubMed] [Google Scholar]
- 17.Morvan F, et al. Deletion of a single allele of the Dkk1 gene leads to an increase in bone formation and bone mass. J Bone Miner Res. 2006;21:934–945. doi: 10.1359/jbmr.060311. [DOI] [PubMed] [Google Scholar]
- 18.Li J, et al. Dkk1-mediated inhibition of Wnt signaling in bone results in osteopenia. Bone. 2006;39:754–766. doi: 10.1016/j.bone.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 19.Glass DA, II, et al. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev Cell. 2005;8:751–764. doi: 10.1016/j.devcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 20.Holmen SL, et al. Essential role of beta-catenin in postnatal bone acquisition. J Biol Chem. 2005;280:21162–21168. doi: 10.1074/jbc.M501900200. [DOI] [PubMed] [Google Scholar]
- 21.Zhang C, et al. Ternary complexes and cooperative interplay between NCoA-62/Ski-interacting protein and steroid receptor coactivators in vitamin D receptor-mediated transcription. J Biol Chem. 2001;276:40614–40620. doi: 10.1074/jbc.M106263200. [DOI] [PubMed] [Google Scholar]
- 22.Lyons JP, et al. Wnt-4 activates the canonical beta-catenin-mediated Wnt pathway and binds Frizzled-6 CRD: Functional implications of Wnt/beta-catenin activity in kidney epithelial cells. Exp Cell Res. 2004;298:369–387. doi: 10.1016/j.yexcr.2004.04.036. [DOI] [PubMed] [Google Scholar]
- 23.Zhang C, et al. Nuclear coactivator-62 kDa/Ski-interacting protein is a nuclear matrix-associated coactivator that may couple vitamin D receptor-mediated transcription and RNA splicing. J Biol Chem. 2001;278:35325–35336. doi: 10.1074/jbc.M305191200. [DOI] [PubMed] [Google Scholar]
- 24.Oosterwegel M, et al. Cloning of murine TCF-1, a T cell-specific transcription factor interacting with functional motifs in the CD3-epsilon and T cell receptor alpha enhancers. J Exp Med. 1991;173:1133–1142. doi: 10.1084/jem.173.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.