Abstract
To reveal regulators of innate immunity, we used RNAi assays to monitor the immune response when genes are inhibited in Caenorhabditis elegans and mouse macrophages. Genes that altered innate immune responsiveness in C. elegans were validated in murine macrophages, resulting in the discovery of 11 genes that regulate the innate immune response in both systems and the subsequent identification of a protein interaction network with a conserved role in innate immunity regulation. We confirmed the role of four of these 11 genes in antimicrobial gene regulation using available mutants in C. elegans. Several of these genes (acy-1, tub-2, and tbc-1) also regulate susceptibility to the pathogen Pseudomonas aeruginosa. These genes may prove critical to understanding host defense and represent potential therapeutic targets for infectious and immunological diseases.
Keywords: Caenorhabditis elegans, macrophage
The innate immune system has substantial effects on the regulation of human disease. Innate immunity initiates the action of phagocytic and cytotoxic cells that are the primary short-term means of defense against infection, making a functional innate immune system critical to host defense (1). However, excessive activation of the innate immune response can contribute to many immunological diseases, including asthma, atherosclerosis, and sepsis (2–4). Polymorphisms that are associated with altered susceptibility to many diseases have been identified in innate immunity genes (3), and several genes in innate immune signaling pathways are under investigation as potential therapeutic targets to modulate the inflammatory response (5–9). Thus, the identification of genes that regulate the innate immune response is critical to both the understanding of immunological disease and to the identification of potential targets for treatment of these diseases.
Although many genes that regulate innate immunity have been identified using genetic; biochemical; and, more recently, genomic approaches (10–19), additional genes and gene networks undoubtedly modulate the innate immune response. To discover regulators of the innate immune response to lipopolysaccharide (LPS), we examined immune function in two model systems, Caenorhabditis elegans and murine macrophages, after inhibition of candidate genes by RNAi. Both assays used a Gram-negative bacterial stimulus to induce the immune response. In the C. elegans assay, we monitored the expression of antimicrobial genes induced by Escherichia coli in the nematode; in the mouse macrophage assay, we monitored E. coli LPS-induced cytokine production in the cell culture system. We examined all of the genes on C. elegans chromosome 1 and 192 candidate genes selected from the literature. The innate immunity genes identified in C. elegans were subsequently examined by using siRNA to inhibit the murine homologous genes in macrophages. This approach led to the discovery of 11 genes and a protein interaction network with conserved roles in the regulation of innate immunity. Because the genes and pathways identified in this study exhibit evolutionarily conserved functions in several model systems, they are likely to be important mediators of the immune response and could be potential therapeutic targets for inflammatory diseases.
Results
RNAi Screen of C. elegans Chromosome 1.
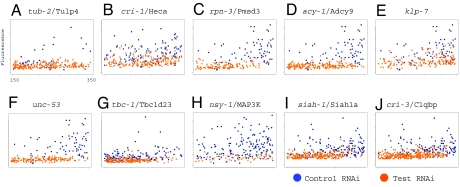
We demonstrated that clec-85::gfp is regulated by multiple immune response pathways in C. elegans in the presence of the nonpathogenic bacterium E. coli and the pathogen Pseudomonas aeruginosa (20). clec-85 expression is also induced by the pathogen Serratia marcescens (21). These observations indicate that clec-85::gfp would be a good screening tool to identify regulators of innate immunity. To identify innate immunity genes, we inhibited each gene on C. elegans chromosome 1, using an E. coli library expressing dsRNA (22), and analyzed changes in clec-85::gfp expression in nematodes harboring a clec-85::gfp transgene (20). Of the 2,416 chromosome 1 genes examined, we identified 32 that regulated clec-85::gfp expression [see Fig. 1 A and B for examples; see supporting information (SI) Table S1 for the full list].
Fig. 1.
Identification of genes that regulate clec-85::gfp expression. Nematodes harboring the clec-85::gfp fusion were treated with the indicated dsRNAs. Depicted are overlay graphs comparing the test RNAi treatments (orange) and a control RNAi treatment (blue); each dot represents a single nematode. The x axis represents the time of flight or length of each animal in COPAS Biosort (Union Biometrica) units, and the y axis represents fluorescence due to the clec-85::gfp fusion. Individual graphs are from several different experiments.
RNAi Screen of C. elegans Orthologs of Candidate Innate Immunity Genes from Other Species.
To investigate the function of previously identified candidate innate immunity genes, the expression of the C. elegans orthologs of these candidates was inhibited and the effect on E. coli-induced clec-85::gfp expression was monitored. E. coli clones expressing dsRNA for 192 genes derived from a literature review were examined, including 38 orthologs of genes induced by LPS in mouse lung tissue, 39 orthologs of Drosophila genes that regulate expression of the antimicrobial gene Diptericin, and several positive- and negative-control genes (Table S2) (10, 14). Also examined were an additional 115 genes potentially involved in immune or stress response pathways (Table S2), including members of MAP kinase, TGF-β, and insulin signaling pathways; orthologs of genes known to regulate mammalian innate immunity; and several genes induced by pathogen exposure in C. elegans (21, 23–27). We identified 25 genes (distinct from the genes on chromosome 1) that were required for full expression of clec-85::gfp (see Fig. 1 C–G for examples, see Table S3 for a full list), including the previously identified immune signaling genes nsy-1 (Fig. 1H) and tir-1 (Table S3) (20, 23, 25, 26).
Although only 4% (5 of 115) of the general stress response pathway genes affected expression of clec-85, 13% (5 of 38) of the orthologs of mouse candidates and 36% (14 of 39) of the orthologs of Drosophila candidates affected nematode antimicrobial gene expression (one additional gene was identified whose sequence did not match the predicted sequence in the RNAi library) (Table S3). In contrast, only 1.3% of genes on chromosome 1 affected clec-85 transcription, suggesting that these candidate genes are highly enriched for potential immune response modulators.
Identification of a Protein Interaction Network That Regulates Innate Immunity.
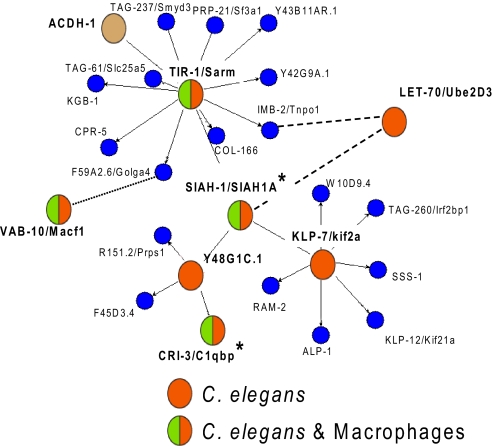
To identify potential protein–protein interactions among our candidate innate immune regulators, we mapped our 57 C. elegans gene products (32 from chromosome 1 and 25 from the candidate screen) onto an interactome that contained the C. elegans interactome database (28) and several manually added orthologous interactions (29–32). Several interaction networks contained a single protein from our screen; however, one protein interaction network contained four proteins from our screen (TIR-1, Y48G1C.1, KLP-7, and LET-70) that directly interacted with a single protein and several other proteins that interacted indirectly (Fig. 2). The four proteins interacted with a common protein, SIAH-1, an E3 ubiquitin ligase that regulates NFκB activity (33) (Fig. 2). One of the SIAH-1 interacting proteins, TIR-1, an ortholog of mammalian Sarm, which is a member of the Myd88 family known to be involved in the nematode and mammalian immune response (23, 26, 34).
Fig. 2.
A protein interaction network with a conserved role in the innate immune response. Depicted is an interactome map derived from http://vidal.dfci.harvard.edu/interactomedb/i-View/interactomeCurrent.pl and modified manually. The map depicts C. elegans protein–protein interactions (solid lines) derived from yeast 2-hybrid data. The dashed lines depict interactions of the orthologous proteins in Drosophila (29, 31, 32); the dotted line depicts an interaction of the orthologous proteins in humans (30). Red circles indicate genes in C. elegans that regulate production of clec-85::gfp. Circles that are red and green are genes that affect both clec-85::gfp production in C. elegans and IL-6 production in macrophages. Blue circles indicate genes that have not been tested. The asterisks indicate two genes that were identified in the interaction network that were subsequently tested in C. elegans and mouse macrophages and that regulate the innate immune response. The brown circle is a gene that controls susceptibility to P. aeruginosa (56).
The SIAH-1 protein interaction network contains several genes that regulate clec-85 transcription, raising the possibility that other genes in the network might also regulate innate immunity. To test this hypothesis, we inhibited two additional genes in the network: siah-1, because it is the central node in the network, and cri-3, because it is homologous to C1qbp, which regulates some aspects of mammalian innate immunity (35). Inhibition of both of these genes caused a decrease in the expression of clec-85::gfp (Fig. 1 I and J). Thus, in addition to the 57 C. elegans genes identified in our chromosome 1 and candidate gene screens, we identified two additional potential innate immune regulators.
Immune Function of Mammalian Orthologs of C. elegans Innate Immunity Genes.
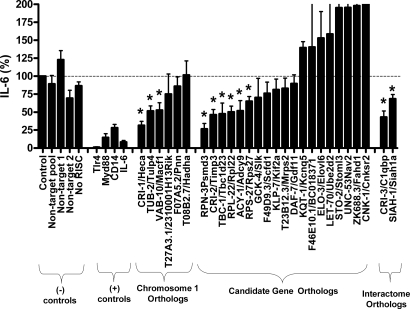
To examine the role of the 59 C. elegans immune response genes in mammalian cells, RNAi was used to inhibit the murine orthologs in a mouse macrophage cell line. Macrophages were transfected with pooled siRNAs (see Materials and Methods), stimulated with E. coli LPS, and then cytokine production was measured. Inhibition of genes known to be involved in the response to LPS, including the LPS receptor Tlr4 and the signaling adaptors Myd88 and CD14, and IL-6 (1, 36, 37), all resulted in decreased production of IL-6. In contrast, negative control siRNA treatments had little or no effect on cytokine production (Fig. 3).
Fig. 3.
Orthologs of C. elegans candidate innate immunity genes regulate the mammalian innate immune response. The indicated siRNA pools were transfected into a mouse macrophage cell line, the cells were induced with E. coli LPS, and IL-6 production was measured. Cytokine production was normalized relative to a negative control siRNA and cell number as described in Materials and Methods and is graphed as the percentage of control IL-6 production. Depicted from left to right are five negative controls (four siRNAs that should not target any gene and one that should not associate with the RISC complex), four positive controls (Tlr4, Myd88, CD14, and IL-6), and 27 test genes orthologous to genes that affect antimicrobial production in C. elegans (C. elegans name listed first, mouse name second). IL-6 values that were significantly different from wild type in the test siRNAs (P < 0.05, t test) are indicated with an asterisk.
Twenty-seven orthologs of the 59 C. elegans genes (6 of 32 genes on chromosome 1, 19 of 25 genes in the candidate library, and 2 of 2 of the genes identified in the interactome) were inhibited with siRNA. Inhibition of 11 of these 27 genes caused a significant decrease in IL-6 production (Fig. 3). Several siRNA treatments that affected IL-6 production also affected production of TNF-α (Fig. S1), further demonstrating the role of these genes in modulating the innate immune response.
To verify that the siRNA effect was specific, we tested multiple individual siRNAs for 9 of these 11 genes. We found that at least two individual siRNAs corresponding to each of these genes inhibited IL-6 production (Fig. S2) and that the siRNAs were inhibiting the expression of the cognate endogenous gene (Fig. S3). Because multiple independent siRNAs for each of these genes induce similar immune phenotypes, our findings demonstrate that 11 genes (3 from C. elegans chromosome 1, 6 from the candidate gene library, and 2 from the protein interaction network) (Table 1) regulate the expression of immune response genes (clec-85 in nematodes and IL-6 and/or TNFα in murine macrophages).
Table 1.
Genes with a conserved role in innate immunity regulation
| Mouse gene | C. elegans gene* | Macrophage† | C. elegans‡ | Drosophila§ | Mouse | Other |
|---|---|---|---|---|---|---|
| Heca | cri-1=K07A1.7 | IL-6, TNF | clec-85 | |||
| Tulp4 | tub-2 = tag-305 | IL-6 | clec-85 P. aeruginosa | Systemic LPS¶ | Expression correlates with other immune response genes (57) | |
| Macf1 | vab-10 | IL-6, TNF | clec-85 | Overexpressed in patients infected with Parvovirus B19 (58) | ||
| Psmd3 | rpn-3 | IL-6, TNF | clec-85 | Diptericin, NFκB | Binds Traf6, NFκB (59) | |
| Timp3 | cri-2 = tag-225 | IL-6 | clec-85 | Regulates cyokines (60,61) | ||
| Tbc1d23 | tbc-1 | IL-6, TNF | clec-85 P. aeruginosa | Lung LPS‖ | ||
| Rpl22 | rpl-22 | IL-6 | clec-85 | Diptericin | Induced in daf-2(−) (62) | |
| Adcy9 | acy-1 | IL-6, TNF | clec-85 P. aeruginosa | Diptericin | cAMP regulates cytokines (39–42) | |
| Rps27 | rps-27 | IL-6 | clec-85 | Induced in daf-2(−) (62) | ||
| C1qbp | cri-3 = F59A2.3 | IL-6, TNF | clec-85 | |||
| Siah1a | siah-1 | IL-6, TNF | clec-85 | Regulates NFκB (33) |
*Because these genes are conserved regulators of innate immunity, we designate the previously unnamed C. elegans genes K07A1.7, tag-225, and F59A2.3 as cri-1, cri-2, and cri-3, respectively, and rename tag-305 as tub-2, for the second C. elegans member of the Tubby family.
†siRNA treatment regulates Il-6 and/or TNF-α production as indicated (this work).
‡RNAi treatment regulates clec-85::gfp expressission . P. aeruginosa indicates that the mutant is susceptible to pathogen (this work).
§Diptericin indicates genes that regulate the LPS-induced production of that antimicrobial in Drosophila; Psmd3 also regulates LPS-induced NFκB nuclear translocation in Drosophila (14).
¶Induced in mouse organs by systemic LPS exposure (I. Yang and D.A.S., unpublished data).
‖Induced in mouse lung tissue by LPS exposure (10).
In Vivo Significance of Innate Immune Genes in C. elegans Mutants.
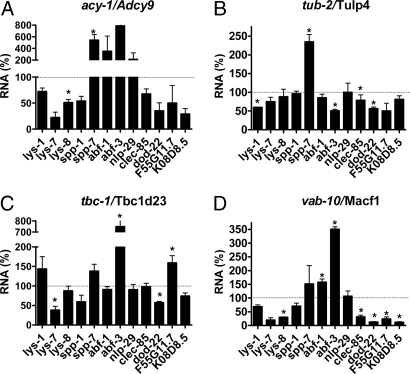
To examine the in vivo significance of these potential innate immune genes, we monitored the production of antimicrobial genes in four available C. elegans mutants (acy-1/Adcy9, tub-2/Tulp4, tbc-1/Tbc1d23, and vab-10/Macf1) of the 11 genes, using qPCR (20) after exposure to E. coli. As observed for known immune response regulators (20), a unique pattern of immune gene expression was observed for each mutant, where deletions both increased and suppressed E. coli-induced gene expression (Fig. 4). Mutations in either acy-1 or vab-10 affected quite a few genes, whereas mutation of tub-2 and tbc-1 had a more subtle effect. These unique antimicrobial gene expression patterns may reflect the complexity of the regulation of the nematode innate immune response.
Fig. 4.
Mutations in candidate innate immune regulators affect expression of many candidate antimicrobial genes. RNA was prepared from the indicated nematode strains and expression of the indicated antimicrobial genes was measured by using qPCR. Gene expression for each mutant is compared to the wild-type strain. Expression values that were significantly different from wild type (P < 0.05, t test) are indicated with an asterisk. Mutant alleles used are: acy-1(md1756), tub-2(gk417), tbc-1(tm2282), and vab-10(e698).
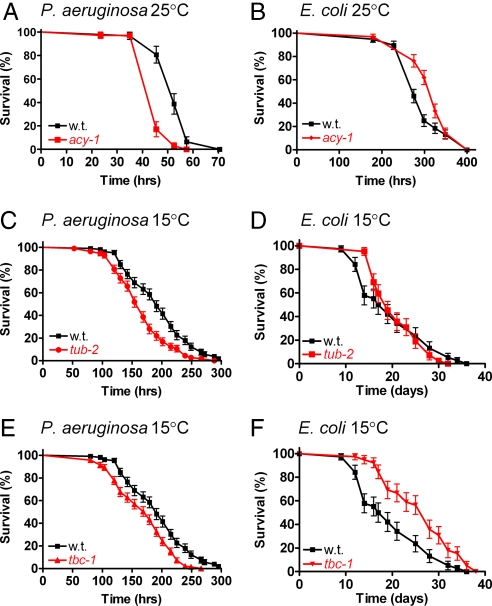
To further examine the in vivo significance of these genes, we tested the response of the four mutant C. elegans strains, acy-1/Adcy9, tub-2/Tulp4, tbc-1/Tbc1d23, and vab-10/Macf1, to the pathogen P. aeruginosa. Nematodes harboring a mutation in acy-1 (an adenylate cyclase) died more rapidly than wild-type nematodes when exposed to P. aeruginosa (Fig. 5A) at 25°C, confirming the requirement for acy-1 in host defense. In contrast, nematodes harboring mutations in tub-2, tbc-1, or vab-10, did not have altered susceptibility to P. aeruginosa at this temperature (data not shown). Kurz et al. (38) demonstrated that small differences in resistance to P. aeruginosa could be better resolved at 15°C; therefore, we monitored the survival of nematodes harboring mutations in either tub-2, tbc-1, or vab-10 in the presence of P. aeruginosa at this lower temperature. Nematodes harboring mutations in either tub-2 or tbc-1 (but not vab-10) were significantly more susceptible to pathogen at this temperature (Fig. 5 C and E). To verify that the short lifespan of these mutant nematodes exposed to P. aeruginosa was due to an innate immune defect and not a general loss of fitness, we examined the survival of acy-1, tub-2, and tbc-1 mutant nematodes in the presence of the nonpathogenic bacterium E. coli. Nematodes harboring these mutations lived as long or slightly longer than wild-type animals in the presence of E. coli (Fig. 2 B, D, and F), indicating that acy-1, tub-2, and tbc-1 likely function in the nematode innate immune response.
Fig. 5.
Identification of genes that regulate C. elegans host defense. Depicted are representative survival plots for either wild-type N2 or the mutants acy-1(md1756), tub-2(gk417), or tbc-1(tm2282). The nematode strains were exposed to the indicated bacteria at the indicated temperature. (A, B, D, and F) Experiments were initiated with 60 nematodes of each strain; a minimum of 36 animals remained through the course of the experiment. (C and E) Experiments were initiated with 200 nematodes; a minimum of 102 animals remained through the course of the experiment. A statistical comparison of the survival plots indicates that survival of the mutant animals was statistically different from the wild-type N2 strain in A (P < 0.0001), B (P = 0.0072), C (P < 0.0001), E (P < 0.00001), and F (P = 0.001) but not in D (P = 0.86).
Discussion
Using comparative genomics, we identified 11 genes and a protein interaction network that regulate innate immunity. Because these genes regulate multiple immune responses in two different model systems, they are likely to be important mediators/modifiers of the innate immune response. Moreover, these genes could represent potential therapeutic targets for infectious and immunological diseases. We were able to verify the in vivo significance of four of these RNAi-identified genes, using available C. elegans mutants; all four mutations altered antimicrobial gene expression, and three of the four affected resistance to a common pathogen, P. aeruginosa.
The Drosophila ortholog of acy-1/Adcy9, one of the innate immune genes discovered by using comparative genomics, regulates the production of an antimicrobial protein, Diptericin (14). Likewise, we found that Adcy9 regulates the production of IL-6 and TNFα in mouse macrophages. This is consistent with previous pharmacological data implicating cAMP in the mammalian cytokine response (39–42). There are nine adenylate cyclases in the mammalian genome; our work identifies Adcy9 as an important regulator of the innate immune response.
The genes tub-2/Tulp4 and tbc-1/Tbc1d23 likewise regulate host defense in the nematode and cytokine production in macrophages. Neither of these genes has been identified as an immune modulator. We also identified seven other genes with conserved innate immune function (Table 1); several of these are likely to also be important innate immune modulators.
Our findings provide further support for the importance of the ubiquitin/26S proteasome pathway in innate immunity (43–45). We identified several genes in the ubiquitin/proteasome pathway, including an E2 ubiquitin ligase (let-70) and several E3 ubiquitin ligases (tag-353, C27A12.6, and siah-1). Consistent with a proposed immune signaling role for these ubiquitin ligases, the mammalian ortholog of tag-353 is thought to associate with the 26S proteasome (46), and Siah1 has been shown to regulate NFκB (33). RPN-3/Psmd3, a component of the regulatory subunit of the 26S proteasome (47), affects the innate immune response in C. elegans, mouse macrophages (this work), and Drosophila (14). Small molecule proteasome inhibitors are undergoing testing for treatment of inflammatory diseases (48–50); our data suggests that siRNA of proteasome subunits may be another potential treatment option.
In mammals, LPS acts through both p38 MAPK and NFκB pathways. A p38 MAPK pathway also functions in C. elegans host defense (25). Although a C. elegans ortholog of NFκB has not been identified (27), our demonstration of the role of the ubiquitin/proteasome pathway in C. elegans host defense suggests that some aspects of this innate immune signaling pathway may be conserved in the nematode. It is possible that the ubiquitin/proteasome system could regulate innate immunity in C. elegans via a different mechanism; alternatively, the pathway could regulate a protein that is highly diverged from NFκB.
Our findings also identify several other genes and a protein interaction network that may be important in controlling the innate immune response. The genes and pathways identified by this comparative genetics approach could be important mediators of innate immunity, and therefore may provide a useful set of targets for treatment of many diseases.
Materials and Methods
RNAi of Candidate Antimicrobial Genes in C. elegans.
RNAi was carried out in liquid culture in 96 well format as described in ref. 20 with specific details outlined in SI Materials and Methods. Nematode fluorescence was assayed by using the COPAS Biosort (Union Biometrica), which measures the length (a measure of nematode age) and fluorescence of each nematode in each well, and analyzed as described in ref. 20.
siRNA in Mouse Macrophages.
To test the role of genes identified in the C. elegans screens on the macrophage immune response, we transfected siRNAs (Dharmacon; pools of four siRNA duplexes per gene) into the mouse macrophage cell line J774A.1 (51). Cells were transfected by using an Amaxa Nucleofector 96-well shuttle, according to the manufacturer's instructions. Transfections were carried out using 2 μM siRNA and 100,000 cells per well. Cells were maintained in DMEM supplemented with 10% FBS. Twenty-four to 36 h after transfection, ultrapure E. coli LPS (List Biological Labs) was added to a final concentration of 20 ng/ml. After 5 h, supernatant was collected, and cytokine production was assayed by using a multiplex cytokine assay kit (Linco) and a Luminex reader (Bio-Rad). Cell viability was monitored, and cell number was normalized by using fluorescien diacetate (52). Cytokine production was normalized relative to a negative control siRNA (53). siRNAs were initially tested in triplicate; siRNAs that prevented IL-6 production were then assayed at least three more times. RNA was isolated and the level of RNA knockdown monitored by qPCR using the primers listed in Table S4.
Assaying the Effect of Candidate Genes on C. elegans Host Defense.
qPCR of available C. elegans mutants was carried out in three independent biological replicates as described in ref. 20, using wild-type N2 as a control.
Survival of C. elegans mutants was monitored at 25°C or 15°C in the presence of P. aeruginosa strain PA14 (54), using N2 as a control. Exposures were initiated when the nematodes were in the late L4 stage. Lifespan analysis, using E. coli strain OP50, was performed in the presence of the sterilizing agent FUDR (55). Survival analysis was carried out by using Graphpad Prism, Version 4.
Supplementary Material
Acknowledgments.
We thank Daniel Snyder and Julie Rice from the National Toxicology Program High-Throughput Screening Core, Shohei Mitani for supplying nematode strains, John Tomfohr for noting the Drosophila LET-70 interactions, Frank Chao for assistance in ortholog identification, and Javier Apfeld for critical reading of the manuscript. This work was supported by the intramural research program of the National Institutes of Health, the National Heart Lung and Blood Institute, the National Institute of Environmental Health Sciences, and the National Toxicology Program. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802405105/DCSupplemental.
References
- 1.Kaufmann SHE, Medzhitov, Gordon . The Innate Immune Response to Infection. Washington, DC: ASM; 2004. [Google Scholar]
- 2.Chaudhuri N, Dower SK, Whyte MK, Sabroe I. Toll-like receptors and chronic lung disease. Clin Sci (London) 2005;109:125–133. doi: 10.1042/CS20050044. [DOI] [PubMed] [Google Scholar]
- 3.Cook DN, Pisetsky DS, Schwartz DA. Toll-like receptors in the pathogenesis of human disease. Nat Immunol. 2004;5:975–979. doi: 10.1038/ni1116. [DOI] [PubMed] [Google Scholar]
- 4.Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 5.Hoebe K, et al. TLR signaling pathways: Opportunities for activation and blockade in pursuit of therapy. Curr Pharm Des. 2006;12:4123–4134. doi: 10.2174/138161206778743466. [DOI] [PubMed] [Google Scholar]
- 6.Hoffman ES, Smith RE, Renaud RC., Jr From the analyst's couch: TLR-targeted therapeutics. Nat Rev Drug Discov. 2005;4:879–880. doi: 10.1038/nrd1880. [DOI] [PubMed] [Google Scholar]
- 7.Ishii KJ, Uematsu S, Akira S. “Toll” gates for future immunotherapy. Curr Pharm Des. 2006;12:4135–4142. doi: 10.2174/138161206778743484. [DOI] [PubMed] [Google Scholar]
- 8.O'Neill LA. Therapeutic targeting of Toll-like receptors for inflammatory and infectious diseases. Curr Opin Pharmacol. 2003;3:396–403. doi: 10.1016/s1471-4892(03)00080-8. [DOI] [PubMed] [Google Scholar]
- 9.Ward SG, O'Neill LA. Spotlight on new anti-inflammatory drug targets in the immune system. Curr Opin Pharmacol. 2003;3:391–395. doi: 10.1016/s1471-4892(03)00085-7. [DOI] [PubMed] [Google Scholar]
- 10.Burch LH, et al. Transcriptional response to endotoxin reveals role for interferon γ in lung neutrophil recruitment. Am J Physiol Lung Cell Mol Physiol. 2006;291:L677–L682. doi: 10.1152/ajplung.00523.2005. [DOI] [PubMed] [Google Scholar]
- 11.Calvano SE, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 12.Cook DN, et al. Genetic regulation of endotoxin-induced airway disease. Genomics. 2004;83:961–969. doi: 10.1016/j.ygeno.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Doyle SL, O'Neill LA. Toll-like receptors: From the discovery of NFkappaB to new insights into transcriptional regulations in innate immunity. Biochem Pharmacol. 2006;72:1102–1113. doi: 10.1016/j.bcp.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Foley E, O'Farrell PH. Functional dissection of an innate immune response by a genome-wide RNAi screen. PLoS Biol. 2004;2:E203. doi: 10.1371/journal.pbio.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoebe K, et al. Genetic analysis of innate immunity. Adv Immunol. 2006;91:175–227. doi: 10.1016/S0065-2776(06)91005-0. [DOI] [PubMed] [Google Scholar]
- 16.Jeyaseelan S, Chu HW, Young SK, Worthen GS. Transcriptional profiling of lipopolysaccharide-induced acute lung injury. Infect Immun. 2004;72:7247–7256. doi: 10.1128/IAI.72.12.7247-7256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemaitre B. The road to Toll. Nat Rev Immunol. 2004;4:521–527. doi: 10.1038/nri1390. [DOI] [PubMed] [Google Scholar]
- 18.O'Neill LA. TLRs: Professor Mechnikov, sit on your hat. Trends Immunol. 2004;25:687–693. doi: 10.1016/j.it.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Yu SL, et al. Differential gene expression in gram-negative and gram-positive sepsis. Am J Respir Crit Care Med. 2004;169:1135–1143. doi: 10.1164/rccm.200211-1278OC. [DOI] [PubMed] [Google Scholar]
- 20.Alper S, McBride SJ, Lackford B, Freedman JH, Schwartz DA. Specificity and complexity of the C. elegans innate immune response. Mol Cell Biol. 2007;27:5544–5553. doi: 10.1128/MCB.02070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mallo GV, et al. Inducible antibacterial defense system in C. elegans. Curr Biol. 2002;12:1209–1214. doi: 10.1016/s0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- 22.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 23.Couillault C, et al. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol. 2004;5:488–494. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- 24.Ewbank JJ The C. elegans Research Community, editor. WormBook. 2006. Signaling in the immune response. Available at www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim DH, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- 26.Liberati NT, et al. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc Natl Acad Sci USA. 2004;101:6593–6598. doi: 10.1073/pnas.0308625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pujol N, et al. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. 2001;11:809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- 28.Li S, et al. A map of the interactome network of the metazoan C elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 30.Kakinuma T, Ichikawa H, Tsukada Y, Nakamura T, Toh BH. Interaction between p230 and MACF1 is associated with transport of a glycosyl phosphatidyl inositol-anchored protein from the Golgi to the cell periphery. Exp Cell Res. 2004;298:388–398. doi: 10.1016/j.yexcr.2004.04.047. [DOI] [PubMed] [Google Scholar]
- 31.Neufeld TP, Tang AH, Rubin GM. A genetic screen to identify components of the sina signaling pathway in Drosophila eye development. Genetics. 1998;148:277–286. doi: 10.1093/genetics/148.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang AH, Neufeld TP, Kwan E, Rubin GM. PHYL acts to down-regulate TTK88, a transcriptional repressor of neuronal cell fates, by a SINA-dependent mechanism. Cell. 1997;90:459–467. doi: 10.1016/s0092-8674(00)80506-1. [DOI] [PubMed] [Google Scholar]
- 33.Polekhina G, et al. Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-alpha signaling. Nat Struct Biol. 2002;9:68–75. doi: 10.1038/nsb743. [DOI] [PubMed] [Google Scholar]
- 34.Carty M, et al. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat Immunol. 2006;7:1074–1081. doi: 10.1038/ni1382. [DOI] [PubMed] [Google Scholar]
- 35.Ghebrehiwet B, Peerschke EI. cC1q-R (calreticulin) and gC1q-R/p33: Ubiquitously expressed multi-ligand binding cellular proteins involved in inflammation and infection. Mol Immunol. 2004;41:173–183. doi: 10.1016/j.molimm.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 36.Akira S. Mammalian Toll-like receptors. Curr Opin Immunol. 2003;15:5–11. doi: 10.1016/s0952-7915(02)00013-4. [DOI] [PubMed] [Google Scholar]
- 37.Takeda K, Akira S. TLR signaling pathways. Semin Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 38.Kurz CL, Shapira M, Chen K, Baillie DL, Tan MW. Caenorhabditis elegans pgp-5 is involved in resistance to bacterial infection and heavy metal and its regulation requires TIR-1 and a p38 map kinase cascade. Biochem Biophys Res Comm. 2007;363:438–443. doi: 10.1016/j.bbrc.2007.08.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aronoff DM, Carstens JK, Chen GH, Toews GB, Peters-Golden M. Short communication: Differences between macrophages and dendritic cells in the cyclic AMP-dependent regulation of lipopolysaccharide-induced cytokine and chemokine synthesis. J Interferon Cytokine Res. 2006;26:827–833. doi: 10.1089/jir.2006.26.827. [DOI] [PubMed] [Google Scholar]
- 40.Bailly S, Ferrua B, Fay M, Gougerot-Pocidalo MA. Differential regulation of IL 6, IL 1 A, IL 1 beta and TNF alpha production in LPS-stimulated human monocytes: Role of cyclic AMP. Cytokine. 1990;2:205–210. doi: 10.1016/1043-4666(90)90017-n. [DOI] [PubMed] [Google Scholar]
- 41.Dahle MK, Myhre AE, Aasen AO, Wang JE. Effects of forskolin on Kupffer cell production of interleukin-10 and tumor necrosis factor alpha differ from those of endogenous adenylyl cyclase activators: Possible role for adenylyl cyclase 9. Infect Immun. 2005;73:7290–7296. doi: 10.1128/IAI.73.11.7290-7296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin CA, Dorf MA. Differential regulation of Interleukin-6, macrophage inflammatory protein-1, and JE/MCP-1 cytokine expression in macrophage cell lines. Cell Immunol. 1991;135:245–258. doi: 10.1016/0008-8749(91)90269-h. [DOI] [PubMed] [Google Scholar]
- 43.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 44.Ben-Neriah Y. Regulatory functions of ubiquitination in the immune system. Nat Immunol. 2002;3:20–26. doi: 10.1038/ni0102-20. [DOI] [PubMed] [Google Scholar]
- 45.Scheidereit C. IkappaB kinase complexes: Gateways to NF-kappaB activation and transcription. Oncogene. 2006;25:6685–6705. doi: 10.1038/sj.onc.1209934. [DOI] [PubMed] [Google Scholar]
- 46.van Laar T, van der Eb AJ, Terleth C. Mif1: A missing link between the unfolded protein response pathway and ER-associated protein degradation? Curr Protein Pept Sci. 2001;2:169–190. doi: 10.2174/1389203013381189. [DOI] [PubMed] [Google Scholar]
- 47.Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 48.Calzado MA, Bacher S, Schmitz ML. NF-kappaB inhibitors for the treatment of inflammatory diseases and cancer. Curr Med Chem. 2007;14:367–376. doi: 10.2174/092986707779941113. [DOI] [PubMed] [Google Scholar]
- 49.Elliott PJ, Zollner TM, Boehncke WH. Proteasome inhibition: A new anti-inflammatory strategy. J Mol Med. 2003;81:235–245. doi: 10.1007/s00109-003-0422-2. [DOI] [PubMed] [Google Scholar]
- 50.Wang J, Maldonado MA. The ubiquitin-proteasome system and its role in inflammatory and autoimmune diseases. Cell Mol Immunol. 2006;3:255–261. [PubMed] [Google Scholar]
- 51.Ralph P, Nakoinz I. Phagocytosis and cytolysis by a macrophage tumour and its cloned cell line. Nature. 1975;257:393–394. doi: 10.1038/257393a0. [DOI] [PubMed] [Google Scholar]
- 52.Fernandez-Botran R, Vťvička V. Methods in Cell Immunology. Boca Raton, FL: CRC; 2001. [Google Scholar]
- 53.Sorensen DR, Leirdal M, Sioud M. Gene silencing by systemic delivery of synthetic siRNAs in adult mice. J Mol Biol. 2003;327:761–766. doi: 10.1016/s0022-2836(03)00181-5. [DOI] [PubMed] [Google Scholar]
- 54.Rahme LG, et al. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 55.Mitchell DH, Stiles JW, Santelli J, Sanadi DR. Synchronous growth and aging of Caenorhabditis elegans in the presence of fluorodeoxyuridine. J Gerontol. 1979;34:28–36. doi: 10.1093/geronj/34.1.28. [DOI] [PubMed] [Google Scholar]
- 56.Shapira M, et al. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc Natl Acad Sci USA. 2006;103:14086–14091. doi: 10.1073/pnas.0603424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Voy BH, et al. Extracting gene networks for low-dose radiation using graph theoretical algorithms. PLoS Comput Biol. 2006;2:e89. doi: 10.1371/journal.pcbi.0020089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kerr JR, et al. Single-nucleotide polymorphisms associated with symptomatic infection and differential human gene expression in healthy seropositive persons each implicate the cytoskeleton, integrin signaling, and oncosuppression in the pathogenesis of human parvovirus B19 infection. J Infect Dis. 2005;192:276–286. doi: 10.1086/430950. [DOI] [PubMed] [Google Scholar]
- 59.Bouwmeester T, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 60.Mohammed FF, et al. Abnormal TNF activity in Timp3−/− mice leads to chronic hepatic inflammation and failure of liver regeneration. Nat Genet. 2004;36:969–977. doi: 10.1038/ng1413. [DOI] [PubMed] [Google Scholar]
- 61.Smookler DS, et al. Tissue inhibitor of metalloproteinase 3 regulates TNF-dependent systemic inflammation. J Immunol. 2006;176:721–725. doi: 10.4049/jimmunol.176.2.721. [DOI] [PubMed] [Google Scholar]
- 62.Halaschek-Wiener J, et al. Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res. 2005;15:603–615. doi: 10.1101/gr.3274805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.