Abstract
Replication forks routinely encounter damaged DNA and tightly bound proteins, leading to fork stalling and inactivation. To complete DNA synthesis, it is necessary to remove fork-blocking lesions and reactivate stalled fork structures, which can occur by multiple mechanisms. To study the mechanisms of stalled fork reactivation, we used a model fork intermediate, the origin fork, which is formed during replication from the bacteriophage T4 origin, ori(34). The origin fork accumulates within the T4 chromosome in a site-specific manner without the need for replication inhibitors or DNA damage. We report here that the origin fork is processed in vivo to generate a regressed fork structure. Furthermore, origin fork regression supports two mechanisms of fork resolution that can potentially lead to fork reactivation. Fork regression generates both a site-specific double-stranded end (DSE) and a Holliday junction. Each of these DNA elements serves as a target for processing by the T4 ATPase/exonuclease complex [gene product (gp) 46/47] and Holliday junction-cleaving enzyme (EndoVII), respectively. In the absence of both gp46 and EndoVII, regressed origin forks are stabilized and persist throughout infection. In the presence of EndoVII, but not gp46, there is significantly less regressed origin fork accumulation apparently due to cleavage of the regressed fork Holliday junction. In the presence of gp46, but not EndoVII, regressed origin fork DSEs are processed by degradation of the DSE and a pathway that includes recombination proteins. Although both mechanisms can occur independently, they may normally function together as a single fork reactivation pathway.
Keywords: 2D gel electrophoresis, DNA replication, recombination, restart
Replication forks routinely encounter various lesions, ranging from damaged nucleotide residues to covalent protein–DNA complexes. The consequences of these encounters depend on the type and position of the lesion (1). Although some lesions result in fork breakage, many result in fork stalling and replisome inactivation. Presumably, replisome disassembly is important for the repair of the original DNA lesion. However, it is then necessary to reactivate stalled forks to complete replication and maintain genome stability. Therefore, it is critical to understand the mechanisms of stalled fork resolution and reactivation.
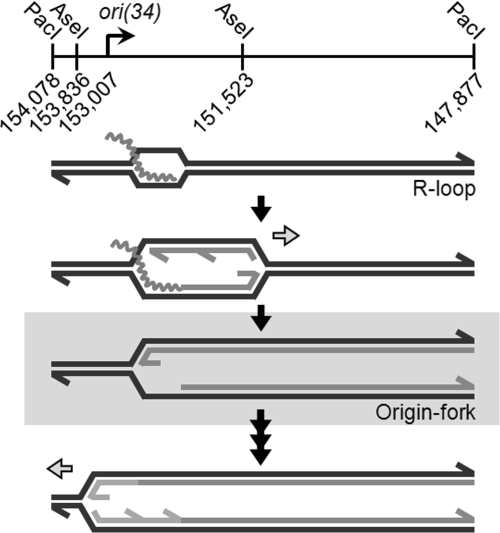
To identify the pathways of stalled fork reactivation, we have used a model fork intermediate that is formed during replication from bacteriophage T4 origin, ori(34). Replication from ori(34) occurs bidirectionally through a two-step process (Fig. 1) (2). A middle-mode promoter and DNA unwinding element promote the formation of an R loop that initiates the first replication fork (3). The mechanism of T4 replication from an R loop has been analyzed in vitro (4). Initially, the branch structure of the R loop supports the binding of the replicative helicase loader [gene product (gp) 59], which promotes removal of the ssDNA-binding protein (gp32) in favor of the replicative helicase (gp41). Gp59 also functions as a gatekeeper, preventing uncoupled leading-strand extension until gp41 is loaded (5, 6). When gp59 is absent, replication proceeds at a slower rate before assembly of the replicative helicase, but the helicase (and the associated primase) does load onto the fork at a reduced efficiency (4). In either case (presence or absence of gp59), when the initial replication fork leaves the origin region, a site-specific branch is left behind at the origin; we refer to this transient fork-shaped intermediate as the origin fork (Fig. 1, gray box). After some delay, the origin fork is converted into a functional retrograde replication fork by unknown mechanisms. Presumably, gp59 could load gp41 directly onto fork intermediates to allow activation without fork processing. However, the delayed activation of the origin fork suggests that other mechanisms also are involved. We propose that the origin fork is a useful model for studying the mechanisms of stalled fork activation in a site-specific manner without the need for replication inhibitors or DNA damage (which typically are used to generate such models in vivo).
Fig. 1.
Replication of T4 origin ori(34). Wavy lines indicate the position of ori(34) transcripts. Horizontal arrows indicate the direction of an active replication fork. The highlighted branched molecule is the origin fork, activation of which (last step) is the subject of this study.
Bacteriophage T4 replication is tightly coupled to recombination. Although replication at early times of infection is origin-dependent, late replication is driven by recombination proteins in a process called recombination-dependent replication (RDR) (7). Gp46/47, a member of the Rad50/Mre11 family, acts on dsDNA ends (DSEs) that arise during T4 infection, promoting end degradation and preparing ends for homologous recombination (8, 9). In this regard, gp46/47 exhibits specific protein–protein interactions with the T4 recombination mediator protein, UvsY (10). This interaction is thought to recruit UvsY to newly generated ssDNA and to promote loading and filamentation of the T4 strand-exchange protein, UvsX. The invasion of a processed DSE into duplex DNA creates a D-loop structure, which, like the R loops discussed in the previous paragraph, can be used to initiate replication. During RDR, the invading 3′ end serves as the primer for leading-strand replication, whereas the displaced ssDNA region facilitates gp59-mediated loading of gp41 (11).
The T4 Holliday junction-cleaving enzyme (EndoVII), the product of gene 49, is a structure-specific endonuclease and was the first enzyme shown to resolve Holliday junctions (12). EndoVII-deficient infections accumulate an abundance of heavily branched intermediates (13). The major function of EndoVII appears to be the resolution of branched molecules before packaging of DNA into phage heads. In contrast to some other resolvases, such as RuvC, EndoVII can act on a broad range of DNA structures in vitro, including fork-shaped structures (14–16).
To analyze the replication and recombination intermediates produced during T4 infection, we have used 2D agarose gel electrophoresis. 2D gels allow separation and visualization of different DNA intermediates based on their size and shape (17). Restriction fragments containing a single replication fork migrate along the Y arc of 2D gels, and the origin fork forms a comet-like shape along the Y arc (2). Recently, several laboratories have reported the appearance of a cone-shaped region that migrates above the Y arc in 2D gels (18–20). The intermediates within this diffuse cone region were assigned as regressed fork intermediates, but this assignment has been challenged. Fierro-Fernandez et al. (21) demonstrated that the 2D gel-migration pattern of simple fork regression does not match the observed cone shape. In addition, several different types of replication intermediates also have been shown to migrate within the cone region (5, 19, 22).
Using 2D agarose gel electrophoresis, we report here that the T4 origin fork undergoes fork regression in vivo and that regression supports two separate mechanisms of origin fork processing. Regression occurs when the newly formed leading and lagging strands of a fork become dissociated from their templates and hybridize to each other (23). This reaction creates a new DSE in the extruded DNA and a Holliday junction at the fork. We find that these two DNA elements serve as targets for processing by gp46/47 and EndoVII, respectively. In addition, we describe the appearance of a modified cone region that results from gp46/47-mediated degradation of the regressed origin fork DSE, explaining how regression can contribute to the formation of a cone region similar to those described previously (18–20).
Results
T4 Origins Provide a Model for Stalled Fork Processing.
Replication of ori(34) involves the formation of a transient fork-shaped intermediate, which we call the origin fork (Fig. 1) (2). Origin forks accumulate at early times of infection and disappear as the infection progresses. Origin replication is repressed at late times by UvsW, an RNA–DNA helicase that displaces the ori(34) transcript from the R loops (24). The disappearance of the origin fork as new initiation is repressed implies processing and resolution of the origin fork.
Several reports have described the appearance of DSEs (and/or broken chromosomes) in association with replication fork stalling (15, 25–28). DSEs could be created from stalled forks by several possible mechanisms, including fork breakage during inactivation, fork cleavage by endonucleases, head-to-tail fork collisions, and fork regression. If the origin fork is a good model for stalled forks, we would expect DSE formation at ori(34).
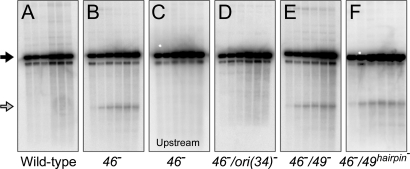
Although we did not detect DSE formation in a wild-type infection, we readily detected a 1.48-kb DSE fragment from the ori(34) region in a 46− mutant infection (after digestion with AseI) (Fig. 2B, open arrow). DSEs are stabilized in 46− infections due to the absence of end processing (29). The ori(34) DSE fragment length is equal to that of one of the newly replicated arms of the origin fork. Furthermore, the 1.48-kb DSE fragment was detected with a probe downstream of ori(34) but not with an upstream probe (Fig. 2 B and C), and the DSE fragment accumulation depends on the presence of a functional ori(34) (Fig. 2D) (30). These results argue strongly that the ori(34) origin fork leads to specific DSE formation. Although the mechanism(s) of DSE formation at ori(34) remain unclear, the appearance of DSE fragments argues that origin forks are subject to the processing pathways invoked for stalled replication forks.
Fig. 2.
Accumulation of DSEs within the ori(34) region. Total DNA from the indicated T4 infections was digested with AseI and separated by agarose gel electrophoresis. DNA was visualized by Southern blot with a probe specific to the DNA region either upstream (C) or downstream (A, B, and D–F) of ori(34). Full-length restriction fragments and the 1.48-kb DSE fragment are indicated by a closed and open arrow, respectively. Note that incomplete digestion of T4-modified DNA results in two full-length linear bands. T4 incorporates 5-hydroxymethylcytosine residues that are further modified with glucosyl groups after replication, which blocks the action of most restriction enzymes.
Accumulation and Resolution of Origin Forks in Various Mutant Infections.
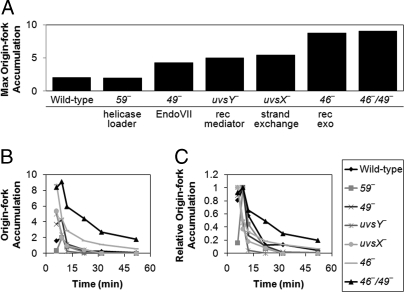
We sought to identify genes that might be involved in the activation of the T4 origin fork. Using 2D agarose gel electrophoresis to visualize replication intermediates, we screened different mutant infections for increased levels of origin fork accumulation. Fig. 3A summarizes the maximum accumulation of origin forks relative to the background level of fork structures present along the Y arc for each of the mutants tested [complete 2D gel time courses can be seen in supporting information (SI) Fig. S1]. Mutational inactivation of several genes reproducibly resulted in a marked increase in maximum origin fork accumulation (46, 49, uvsX, and uvsY). However, the accumulation of origin forks would depend on their rates of formation and resolution. Therefore, any one of these mutations could potentially be increasing origin fork formation (e.g., by stabilizing the R loop) or decreasing fork resolution. Interestingly, mutation of gene 59 did not result in an increase of origin forks during infection. The simplest interpretation is that direct loading of gp41 by gp59 is not a major pathway of origin fork activation.
Fig. 3.
Accumulation and resolution of origin forks. Total DNA from the indicated T4 infections was digested with PacI and separated by 2D gel electrophoresis. Gels were visualized by Southern blot with a probe specific to the ori(34) region. The level of origin forks in each 2D gel is expressed as the fold excess of origin forks relative to the background level of fork structures along the Y arc (measured at a site adjacent to the origin fork). The complete 2D gel time courses are shown in Fig. S1. (A) Maximum origin fork accumulation from each phage infection (not always the same time point). (B) Origin fork accumulation throughout time course of the infections. (C) Origin fork accumulation throughout infection, but with each curve normalized to one at the maximum accumulation. For each affected mutant, a relative increase at the maximal time point was seen in at least one additional experimental replicate. We also quantitated three repeats of the wild-type and five repeats of the 46−/49− double mutant, finding the following values (average ± standard deviations): 2.46 ± 0.84 (wild type) and 8.77 ± 1.63 (46−/49−). The functions of the T4 proteins are indicated in A. gp59, replicative helicase loading protein; gp49, EndoVII (cleaves branched DNA); UvsY, recombination mediator (loads UvsX); UvsX, strand exchange protein; gp46, component of the gp46/47 recombination exonuclease (Rad50/Mre11homolog).
We next asked whether any of the mutants show a deficiency in origin fork resolution over time by estimating the intensity of origin forks at different times of infection (Fig. 3B and Fig. S1). We also normalized the time courses for maximum accumulation to allow a direct comparison of the different infections (Fig. 3C and Fig. S1). Despite the increased accumulation seen in several mutants, only one phage strain, the double mutant 46−/49−, showed a striking delay in origin fork resolution. Interestingly, both 46 and 49 single-mutant infections displayed near wild-type rates of resolution. These results indicate that the products of genes 46 and 49 can resolve origin forks through two separate mechanisms and that either mechanism is sufficient for wild-type rates of resolution.
Regressed Fork Intermediates Are Stabilized in 46−/49− Infections.
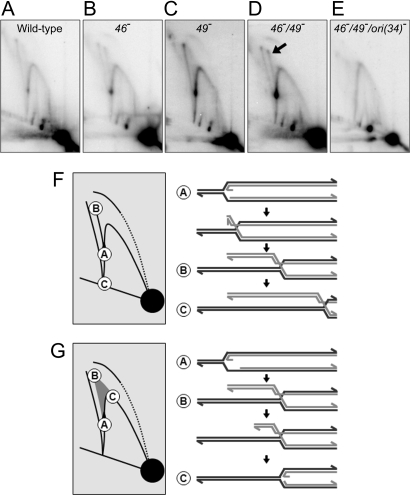
The double 46−/49− mutant infections showed a striking accumulation of a replication intermediate that was barely detectable in wild-type or single-mutant infections; this intermediate is indicated by the arrow in Fig. 4D (also see Fig. S1). The intermediate formed an arc with a migration pattern similar to that seen for X-shaped molecules, which consist of two full-length restriction fragments connected by a Holliday junction. However, the molecules in the arc are smaller in total size than X-shaped molecules, and the arc appears to originate from the origin fork position. The arc is very similar to arcs described by Fierro-Fernandez et al. (21), which were shown to contain regressed replication fork molecules (see Fig. 4F).
Fig. 4.
Replication intermediates from mutant infections. (A–E) The 2D gel image with maximum origin fork accumulation (9 min after infection) is shown for each phage infection indicated. Complete 2D gel time courses are shown in Fig. S1. (F and G) The 2D gel diagrams show the migration of origin forks undergoing regression (F) and regressed origin forks undergoing DSE degradation (G). The structures of different intermediates are shown with the corresponding 2D gel migration position indicated.
The plasmid-based model fork analyzed by Fierro-Fernandez et al. (21) was shown to undergo regression in vitro after restriction enzyme digestion. However, we found that the accumulation of the intermediate was specific to particular infections (e.g., 46−/49− infections), whereas the origin fork accumulated without the arc in various other infections (Fig. S1). These results argue that the putative regressed forks are forming in vivo in the 46−/49− infections, not during extraction or DNA processing. To test this inference more directly, we analyzed DNA from a 46−/49− infection that had undergone interstrand cross-linking before cell lysis and DNA preparation (Fig. S2 C and D). Cross-linking was confirmed by heat-treating a portion of the sample to 100°C for 5 min, followed immediately by electrophoresis (Fig. S2 B and D). Most of the cross-linked DNA migrated as duplex (i.e., had rapidly renatured because of the cross-links). Moreover, the melted and rapidly renatured DNA still showed the arc, indicating that all of the arms had been cross-linked in vivo. These results strongly argue that fork regression occurred in vivo.
If the intermediate is formed by origin fork regression, then it should only be detected with a functional ori(34). We therefore introduced an ori(34) mutation (30) into the 46−/49− phage background. As predicted, the 46−/49−/ori(34)− phage infections did not lead to the appearance of regressed fork molecules (Fig. 4E).
As a final test for the structure of the intermediate, we used a variant of 2D gel electrophoresis that allows some spontaneous branch migration in between the first and second dimensions (21). Extensive branch migration of a regressed fork can result in complete extrusion of the regressed arm and generation of the full-length restriction fragment. The resulting 2D gel of DNA from 46−/49− infections (9 min after infection) showed the loss of both X-shaped molecules (which contain Holliday junctions) and the regressed fork intermediate, whereas fork-shaped molecules (including the origin fork) were not resolved (Fig. S3). This result was accompanied by the appearance of full-length linear fragments and a spot that migrates at the expected position for an extruded 5.2-kb regressed arm (Fig. S3, dotted circle). Taken together, these results argue strongly that origin forks are processed into regressed fork intermediates in vivo, which are stabilized by mutational inactivation of gp46 and gp49.
The Role of EndoVII in Regressed Fork Cleavage.
In the presence of EndoVII, but not gp46 (46−/49+ infections), we detected ≈2-fold less regressed origin forks (as a fraction of total replication intermediates) compared with 46−/49− infections (compare Fig. 4 B and D). This decrease is consistent with EndoVII-mediated cleavage of the regressed fork Holliday junction. However, previous reports have suggested that EndoVII also cleaves Y-shaped molecules, such as the origin fork, in vivo (14, 15). Direct cleavage of the origin fork junction by EndoVII would decrease the number of origin forks available for regression, indirectly decreasing the accumulation of regressed origin forks. Contrary to this theory, we found nearly identical amounts of origin forks (as a fraction of total replication intermediates) from the 46− and 46−/49− infections (Fig. 3A). We also analyzed the cleavage products produced during these infections. Direct cleavage of the origin fork junction by EndoVII would produce DSE fragments of the same size as those seen in Fig. 2 (1.48 kb). However, the same size fragment also could be generated by branch migration of the Holliday junction in the regressed fork past the restriction enzyme site regardless of whether the junction is cleaved. We detected a very modest decrease in the generation of the 1.48-kb band in the 46−/49− infection compared with the 46− infection (average of ≈15% decrease), but this small decrease was not statistically significant. Particularly because the 1.48-kb fragment can be potentially generated by multiple pathways, further analysis is required to determine which origin fork structure(s) are cleaved by EndoVII in vivo.
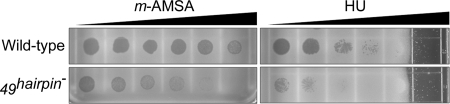
To further investigate the possible role of EndoVII in fork processing, we created a gene 49hairpin mutation. The 49hairpin mutation disrupts the formation of a hairpin on the gene 49 transcript that is believed to prevent expression of full-length EndoVII at early times of infection (31). The 49hairpin mutation should therefore result in unusually early expression of EndoVII during infection. The 49hairpin mutation dramatically increases sensitivity to two different inhibitors that block replication forks: 4′-(9-acridinylamino) methanesulfon-m-anisidide (m-AMSA) and hydroxyurea (HU) (Fig. 5). These inhibitors block fork progression by inducing topoisomerase cleavage complexes and limiting deoxynucleotide production, respectively (32, 33). The hypersensitivity phenotypes suggest that stalled fork intermediates or their byproducts are cleaved by EndoVII when it is expressed at early times of infection. We also introduced the 49hairpin mutation into a 46− background to see whether it would lead to increased production of the ori(34) DSE fragment but failed to find a significant change (Fig. 2F).
Fig. 5.
49hairpin mutation causes hypersensitivity to m-AMSA and HU. Lawns of E. coli MCS1 were prepared on square Petri dishes with a gradient of m-AMSA ≤75 μg/ml or HU ≤2.25 mg/ml (direction of concentration gradient indicated at top). Aliquots of ≈300 plaque-forming units of wild-type or 49hairpin− phage were spotted six times across the gradient. Note that growth of the MCS1 cells is prevented at the highest HU concentrations.
Taken together, these results suggest that EndoVII cleaves, at most, a limited amount of origin forks directly in vivo. Nevertheless, the hypersensitivity phenotypes of the 49hairpin mutant and the decrease in regressed origin forks in EndoVII-proficient infections suggest that EndoVII cleaves the Holliday junction of regressed forks in vivo.
Gp46/47 and Regressed Fork Degradation.
In the presence of gp46, but not EndoVII (46+/49− infections), the regressed fork arc is absent. Nonetheless, we detect a transient accumulation of intermediates that form a modified cone shape, rather than a discrete arc (compare Fig. 4 C and D). The modified cone region forms a heavy smear that extends from the regressed arc position down to the Y arc. This pattern is consistent with gp46/47-mediated degradation of the DSE produced by origin fork regression (see Fig. 4G).
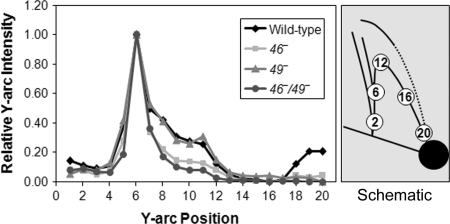
According to this interpretation, the cone region consists of intermediates that have undergone differing amounts of branch migration and DSE degradation. If the regressed DSE is completely degraded by gp46/47, then it should produce a simple Y-shaped molecule that is smaller in total size than the origin fork (i.e., migrating above and to the right of the origin fork along the Y arc). Consistent with this prediction, we detected an increased accumulation in this region of the Y arc in 46+ infections (both wild-type and 49− infections) (Fig. 6). The simplest interpretation is that regressed DSE degradation is responsible for this increased Y-arc accumulation. However, we cannot rule out an alternate possibility (i.e., that the leading and lagging strands of the origin fork are degraded without regression by some gp46/47-dependent process).
Fig. 6.
Accumulation of fork-shaped molecules along the Y arc of 2D gels during infection. The intensity of the Y arc was measured in progressive circles for the 2D gels of wild-type, 46−, 49−, and 46−/49− infections (from Fig. 4 A–D, respectively). To compare fork accumulation among gels, values were normalized for maximum intensity at the position of the unprocessed origin fork.
Discussion
The phage T4 ori(34) origin fork provides a useful model for studying the mechanisms of replication fork reactivation. The position of the origin fork is set by the origin sequence, which generates a site-specific fork intermediate that is formed naturally without the use of replication inhibitors or DNA damage. However, it is important to stress that the origin fork is not a bona fide stalled replication fork, and these studies analyze fork activation rather than reactivation. This study does not address the immediate consequences of fork stalling per se on the replication machinery or stalled fork structure. Instead, the uniform nature of the origin fork allows for focused analysis of the mechanisms that contribute to fork processing and activation.
Although T4 DNA replication is origin-dependent at the earliest times of infection, late replication occurs in a recombination-dependent fashion. Our results argue that even the early intermediates of replication at the T4 origins can be processed by recombination proteins, including gp46/47 and EndoVII. Although some origin forks might be activated directly by gp59-dependent loading of the replisome (6), the persistence of origin forks in the 46−/49− double mutant argues that a significant fraction are normally processed by recombination proteins. Given the tight link between T4 replication and recombination, it is perhaps not surprising that recombination and regression play a prominent role in the activation of stationary fork structures at T4 origins.
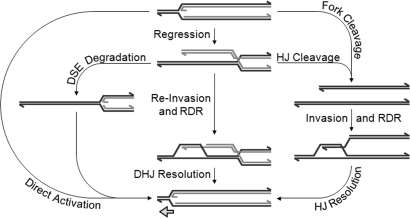
A number of fork reactivation models have invoked a regressed fork intermediate (1). Fork regression involves the formation of a DSE and a Holliday junction, and we have shown here that these two DNA elements serve as targets for resolution by gp46/47 and EndoVII, respectively. When both mechanisms of resolution are disabled (during 46−/49− infections), regressed origin forks are stabilized throughout infection, suggesting that regression is a dominant pathway of fork reactivation in T4. Although 46−/49− infections accumulate a large population of regressed origin forks, a limited accumulation also is seen in wild-type infections at early times (Fig. 4A and Fig. S1). Therefore, regressed forks are formed naturally during wild-type infections and are merely stabilized by gene 46 and 49 mutations. Although the mechanisms of fork resolution supported by gp46/47 and EndoVII can occur independently of each other (46−/49+ and 46+/49− infections), both proteins might normally function together in a single recombination-dependent fork reactivation pathway (see Fig. 7).
Fig. 7.
Pathways of fork processing and activation. Holliday and double-Holliday junctions are abbreviated HJ and DHJ, respectively. In the T4 system, EndoVII could catalyze fork cleavage and HJ/DHJ cleavage/resolution, UvsX catalyzes the strand invasion or reinvasion (assisted by UvsY), and the gp46/47 complex participates in DSE degradation. Direct fork activation could potentially be promoted by the helicase-loader gp59, and the mechanism of fork regression is still unknown.
Degradation of a regressed DSE by gp46/47 may be coupled to strand invasion and homologous recombination because gp46/47 prepares broken ends for strand invasion (9). Formation of a D loop could thereby trigger a localized RDR event near the origin, which would indeed represent a simple mechanism of origin fork activation. Because T4 infections can involve numerous chromosome copies from both coinfection and previous rounds of replication, DSE invasion could in theory favor either intra- or intermolecular recombination. Particularly if Holliday junction cleavage (at the regressed fork) is delayed until after strand invasion, then the regressed DSE may favor intramolecular recombination (i.e., formation of a D loop just ahead of the regressed fork). The ori(34) region is not a recombination hotspot when the DNA is undamaged (30), arguing against high levels of intermolecular DSE invasion.
Previous Escherichia coli studies suggest coordination between fork regression and Holliday junction cleavage (34). Thus, Holliday junction cleavage might be directly influenced by interactions with factors that promote fork regression. In addition, Donaldson et al. (35) have proposed that fork regression and DSE degradation are coordinated in E. coli. In their model, degradation of the regressed duplex is coupled to regression, which limits its length and likely discourages intermolecular recombination.
Several laboratories have recently reported the appearance of a cone-shaped region in 2D gels. Although this collection of intermediates has been attributed as regressed forks, Fierro-Fernandez et al. (21) showed that the migration pattern expected from simple regression differs from that of the cone shapes reported (also see the Introduction). Our data provide a possible explanation for this discrepancy: The cone shapes reported recently are consistent with regressed fork intermediates that have undergone DSE degradation (Fig. 4 C and G).
Many of the T4 proteins analyzed in this study have both bacterial and eukaryotic counterparts that could serve similar functions following fork regression in other systems. In E. coli, both RusA (36) and RuvC (37) are Holliday junction-cleaving enzymes, and RuvC has been proposed to cleave Holliday junctions generated by fork regression (38). In Schizosaccharomyces pombe and Saccharomyces cerevisiae, Mus81 complexes have recently been shown to promote Holliday junction resolution in vitro (39), although their role in vivo is less understood. As mentioned earlier, gp46/47 is a member of the Rad50/Mre11 family, which includes SbcCD in E. coli. Gp46/47 also shares functional similarities with RecBCD (40) in E. coli. All three protein complexes appear to play important roles in DNA processing dependent on the target DNA structure. Interestingly, Rad50/Mre11 has been shown to interact with stalled replication forks (41) and could well play a similar role in processing DSEs generated by fork regression.
Materials and Methods
Bacterial Phage and Strains.
The bacterial host for phage infections is a derivative of E. coli CAG12135 with the following additional mutations: acrA::Tn10-kan, recA::Tn10-cam, and recD. E. coli strain MCS1 (42) was used to produce a lawn on the gradient plates. The phage T4 strains used in this study are all derivatives of the K10 strain [amB262 (gene 38), amS29 (gene 51), nd28 (denA), and rIIPT8 (rII-denB deletion)], which is considered wild type for this study (43). The additional phage T4 mutations used in this study include different combinations of amB14 (gene 46), amE727 (gene 49), amHL628 (gene 59), am11 (uvsX), uvsYΔ (also referred to as K10-608) (42), gene 49hairpin− (GG→TT mutation at T4 coordinates 46880–46881), and ori(34)− (six-point mutations at T4 coordinates 153017–153035) (30).
T4 Infection and DNA Preparation.
Bacterial cells were grown with vigorous shaking at 37°C to a cell density of 4 × 108 per ml (OD560 = 0.5) and infected with the indicated phage strain at a multiplicity of six plaque-forming units per cell. After 4 min at 37°C without shaking (for adsorption), incubation was continued with shaking for the indicated time. For each time point, infected cells from 1 ml of culture were collected by centrifugation at 15,800 × g for 2 min and resuspended in 300 μl of lysis buffer [50 mM Tris·HCl (pH 7.8), 10 mM disodium EDTA, 100 mM NaCl, and 0.2% SDS]. Proteinase K was added to 0.5 mg/ml, and the resuspension was incubated at 65°C for 2 h. Total nucleic acid was extracted sequentially with phenol and chloroform-isoamyl alcohol (24:1) and then dialyzed overnight at 4°C against TE buffer [10 mM Tris·HCl (pH 7.8) and 1 mM disodium EDTA].
2D Agarose Gel Electrophoresis.
Total DNA from infected cells was treated with restriction endonuclease PacI (New England Biolabs) before analysis by 2D agarose gel electrophoresis according to Friedman and Brewer (17). Additional details about 2D gel electrophoresis conditions, including the modified extrusion method, are found in Figs. S1 and S3.
Southern Blots.
Agarose gels were transferred to a 0.45-μm Nytran transfer membrane (Whatman) by the downward sponge method (44). After transfer, the DNA was cross-linked to the membrane with a 120 mJ/cm2 UV exposure. Radiolabeled probes were generated by using a PCR fragment and the Random-Primed DNA Labeling kit (Roche). All blots were visualized by using the Storm 860 PhosphorImager (Molecular Dynamics) and quantitated with ImageQuant software (Molecular Dynamics).
DSE Accumulation Assay.
Total DNA from infection was treated with restriction endonuclease AseI (New England Biolabs) before separation on a 1% agarose gel run in 0.5× TBE buffer for 12 h at 2 V/cm at room temperature. The gel was visualized by Southern blot with a probe that is specific to a region either upstream (T4 coordinates 153195–153783) or downstream (T4 coordinates 151767–152688) of ori(34). Quantitation of DSE accumulation is expressed as a fraction of total linear molecules present at each time point.
Drug-Sensitivity Gradient Plates.
Phage sensitivity to m-AMSA and HU was analyzed by using gradient plates as described by Woodworth and Kreuzer (45). First, 25 ml of T4 agar (13 g of Bactotryptone, 8 g of NaCl, 2 g of sodium citrate dihydrate, 1.3 g of glucose, and 10 g of agar per liter) with 75 μg/ml m-AMSA or 2.25 mg/ml HU was poured into a square Petri plate with one edge elevated. Once solidified, the plate was leveled and another 25 ml of T4 agar (without drug) was added. A lawn of E. coli was produced by growing MCS1 to a cell density of 4 × 108 per ml and then plating 500 μl of cells in 5 ml of drug-free T4 top agar (same as T4 agar except with 4 g of agar per liter). Aliquots of ≈300 T4 plaque-forming units were then spotted across the gradient plate.
Supplementary Material
Acknowledgments.
This research was supported by National Institutes of Health Grant GM 072089 (to K.N.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711999105/DCSupplemental.
References
- 1.McGlynn P, Lloyd RG. Recombinational repair and restart of damaged replication forks. Nat Rev Mol Cell Biol. 2002;3:859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- 2.Belanger K, Kreuzer KN. Bacteriophage T4 initiates bidirectional DNA replication through a two-step process. Mol Cell. 1998;2:693–701. doi: 10.1016/s1097-2765(00)80167-7. [DOI] [PubMed] [Google Scholar]
- 3.Carles-Kinch K, Kreuzer KN. RNA-DNA hybrid formation at a bacteriophage T4 replication origin. J Mol Biol. 1997;266:915–926. doi: 10.1006/jmbi.1996.0844. [DOI] [PubMed] [Google Scholar]
- 4.Nossal NG, Dudas KC, Kreuzer KN. Bacteriophage T4 proteins replicate plasmids with a preformed R Loop at the T4 ori(uvsY) replication origin in vitro. Mol Cell. 2001;7:31–41. doi: 10.1016/s1097-2765(01)00152-6. [DOI] [PubMed] [Google Scholar]
- 5.Dudas KC, Kreuzer KN. Bacteriophage T4 helicase loader protein gp59 functions as gatekeeper in origin-dependent replication in vivo. J Biol Chem. 2005;280:21561–21569. doi: 10.1074/jbc.M502351200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xi J, et al. Interaction between the T4 helicase-loading protein (gp59) and the DNA polymerase (gp43): A locking mechanism to delay replication during replisome assembly. Biochemistry. 2005;44:2305–2318. doi: 10.1021/bi0479508. [DOI] [PubMed] [Google Scholar]
- 7.Mosig G. Relationship of T4 DNA replication and recombination. In: Mathews CK, Kutter EM, Mosig G, Berget PB, editors. Bacteriophage T4. Washington, DC: American Society for Microbiology; 1983. pp. 120–130. [Google Scholar]
- 8.Mickelson C, Wiberg J. Membrane-associated DNase activity controlled by genes 46 and 47 of bacteriophage T4D and elevated DNase activity associated with the T4 das mutation. J Virol. 1981;40:65–77. doi: 10.1128/jvi.40.1.65-77.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cromie GA, Connelly JC, Leach DRF. Recombination at double-strand breaks and DNA ends: Conserved mechanisms from phage to humans. Mol Cell. 2001;8:1163–1174. doi: 10.1016/s1097-2765(01)00419-1. [DOI] [PubMed] [Google Scholar]
- 10.Yassa D, Chou K, Morrical S. Characterization of an amino-terminal fragment of the bacteriophage T4 uvsY recombination protein. Biochimie. 1997;79:275–285. doi: 10.1016/s0300-9084(97)83515-8. [DOI] [PubMed] [Google Scholar]
- 11.Morrical SW, Beernink HTH, Dash A, Hempstead K. The gene 59 protein of bacteriophage T4. Characterization of protein–protein interactions with gene 32 protein, the T4 single-stranded DNA binding protein. J Biol Chem. 1996;271:20198–20207. doi: 10.1074/jbc.271.33.20198. [DOI] [PubMed] [Google Scholar]
- 12.Mizuuchi K, Kemper B, Hays J, Weisberg R. T4 endonuclease VII cleaves Holliday structures. Cell. 1982;29:357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- 13.Kemper B, Brown D. Function of gene 49 of bacteriophage T4. II. Analysis of intracellular development and the structure of very fast-sedimenting DNA. J Virol. 1976;18:1000–1015. doi: 10.1128/jvi.18.3.1000-1015.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensch F, Kemper B. Endonuclease VII resolves Y-junctions in branched DNA in vitro. EMBO J. 1986;5:181–189. doi: 10.1002/j.1460-2075.1986.tb04194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong G, Kreuzer KN. Endonuclease cleavage of blocked replication forks: An indirect pathway of DNA damage from antitumor drug-topoisomerase complexes. Proc Natl Acad Sci USA. 2003;100:5046–5051. doi: 10.1073/pnas.0835166100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruber M, Wellinger RE, Sogo JM. Architecture of the replication fork stalled at the 3′ end of yeast ribosomal genes. Mol Cell Biol. 2000;20:5777–5787. doi: 10.1128/mcb.20.15.5777-5787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman K, Brewer B. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 1995;262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- 18.Courcelle J, Donaldson JR, Chow K-H, Courcelle CT. DNA damage-induced replication fork regression and processing in Escherichia coli. Science. 2003;299:1064–1067. doi: 10.1126/science.1081328. [DOI] [PubMed] [Google Scholar]
- 19.Lopes M, et al. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 20.Vengrova S, Dalgaard JZ. RNase-sensitive DNA modification(s) initiates S. pombe mating-type switching. Genes Dev. 2004;18:794–804. doi: 10.1101/gad.289404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fierro-Fernandez M, Hernandez P, Krimer DB, Schvartzman JB. Replication fork reversal occurs spontaneously after digestion but is constrained in supercoiled domains. J Biol Chem. 2007;282:18190–18196. doi: 10.1074/jbc.M701559200. [DOI] [PubMed] [Google Scholar]
- 22.Martin-Parras L, Hernandez P, Martinez-Robles ML, Schvartzman JB. Unidirectional replication as visualized by two-dimensional agarose gel electrophoresis. J Mol Biol. 1991;220:843–853. doi: 10.1016/0022-2836(91)90357-c. [DOI] [PubMed] [Google Scholar]
- 23.Higgins NP, Kato K, Strauss B. A model for replication repair in mammalian cells. J Mol Biol. 1976;101:417–425. doi: 10.1016/0022-2836(76)90156-x. [DOI] [PubMed] [Google Scholar]
- 24.Dudas KC, Kreuzer KN. UvsW protein regulates bacteriophage T4 origin-dependent replication by unwinding R-loops. Mol Cell Biol. 2001;21:2706–2715. doi: 10.1128/MCB.21.8.2706-2715.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davidson IF, Li A, Blow JJ. Deregulated replication licensing causes DNA fragmentation consistent with head-to-tail fork collision. Mol Cell. 2006;24:433–443. doi: 10.1016/j.molcel.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuzminov A. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc Natl Acad Sci USA. 2001;98:8241–8246. doi: 10.1073/pnas.131009198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michel B, Ehrlich SD, Uzest M. DNA double-strand breaks caused by replication arrest. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seigneur M, Ehrlich SD, Michel B. RuvABC-dependent double-strand breaks in dnaBts mutants require RecA. Mol Microbiol. 2000;38:565–574. doi: 10.1046/j.1365-2958.2000.02152.x. [DOI] [PubMed] [Google Scholar]
- 29.Kreuzer K, Saunders M, Weislo L, Kreuzer H. Recombination-dependent DNA replication stimulated by double-strand breaks in bacteriophage T4. J Bacteriol. 1995;177:6844–6853. doi: 10.1128/jb.177.23.6844-6853.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doan PL, Belanger KG, Kreuzer KN. Two types of recombination hotspots in bacteriophage T4: One requires DNA damage and a replication origin and the other does not. Genetics. 2001;157:1077–1087. doi: 10.1093/genetics/157.3.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosig G, Luder A, Ernst A, Canan N. Bypass of a primase requirement for bacteriophage T4 DNA replication in vivo by a recombination enzyme, endonuclease VII. New Biologist. 1991;3:1195–1205. [PubMed] [Google Scholar]
- 32.Hong G, Kreuzer KN. An antitumor drug-induced topoisomerase cleavage complex blocks a bacteriophage T4 replication fork in vivo. Mol Cell Biol. 2000;20:594–603. doi: 10.1128/mcb.20.2.594-603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noguchi E, Noguchi C, Du L-L, Russell P. Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol Cell Biol. 2003;23:7861–7874. doi: 10.1128/MCB.23.21.7861-7874.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGlynn P, Lloyd RG. Action of RuvAB at replication fork structures. J Biol Chem. 2001;276:41938–41944. doi: 10.1074/jbc.M107945200. [DOI] [PubMed] [Google Scholar]
- 35.Donaldson JR, Courcelle CT, Courcelle J. RuvABC is required to resolve Holliday junctions that accumulate following replication on damaged templates in Escherichia coli. J Biol Chem. 2006;281:28811–28821. doi: 10.1074/jbc.M603933200. [DOI] [PubMed] [Google Scholar]
- 36.Chan SN, Harris L, Bolt EL, Whitby MC, Lloyd RG. Sequence specificity and biochemical characterization of the RusA Holliday junction resolvase of Escherichia coli. J Biol Chem. 1997;272:14873–14882. doi: 10.1074/jbc.272.23.14873. [DOI] [PubMed] [Google Scholar]
- 37.Bennett R, Dunderdale H, West S. Resolution of Holliday junctions by RuvC resolvase: Cleavage specificity and DNA distortion. Cell. 1993;74:1021–1031. doi: 10.1016/0092-8674(93)90724-5. [DOI] [PubMed] [Google Scholar]
- 38.Seigneur M, Bidnenko V, Ehrlich S, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 39.Gaskell LJ, Osman F, Gilbert RJC, Whitby MC. Mus81 cleavage of Holliday junctions: A failsafe for processing meiotic recombination intermediates? EMBO J. 2007;26:1891–1901. doi: 10.1038/sj.emboj.7601645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bleuit JS, et al. Mediator proteins orchestrate enzyme-ssDNA assembly during T4 recombination-dependent DNA replication and repair. Proc Natl Acad Sci USA. 2001;98:8298–8305. doi: 10.1073/pnas.131007498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robison JG, Elliott J, Dixon K, Oakley GG. Replication protein A and the Mre11-Rad50-Nbs1 complex co-localize and interact at sites of stalled replication forks. J Biol Chem. 2004;279:34802–34810. doi: 10.1074/jbc.M404750200. [DOI] [PubMed] [Google Scholar]
- 42.Kreuzer K, Engman H, Yap W. Tertiary initiation of replication in bacteriophage T4. Deletion of the overlapping uvsY promoter/replication origin from the phage genome. J Biol Chem. 1988;263:11348–11357. [PubMed] [Google Scholar]
- 43.Selick H, Kreuzer K, Alberts B. The bacteriophage T4 insertion/substitution vector system. A method for introducing site-specific mutations into the virus chromosome. J Biol Chem. 1988;263:11336–11347. [PubMed] [Google Scholar]
- 44.Ming Y, Di X, Gomez-Sanchez E, Gomez-Sanchez C. Improved downward capillary transfer for blotting of DNA and RNA. Biotechniques. 1994;16:58–59. [PubMed] [Google Scholar]
- 45.Woodworth DL, Kreuzer KN. Bacteriophage T4 mutants hypersensitive to an antitumor agent that induces topoisomerase-DNA cleavage complexes. Genetics. 1996;143:1081–1090. doi: 10.1093/genetics/143.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.