Abstract
Cell surface-located nucleoside triphosphate diphosphohydrolases (NTPDase1, -2, -3, and -8) are oligomeric integral membrane proteins responsible for signal conversion and inactivation in extracellular nucleotide-mediated “purinergic” signaling. They catalyze the sequential hydrolysis of the signaling molecule ATP via ADP to AMP. Here we present the structure of the extracellular domain of Rattus norvegicus NTPDase2 in an active state at resolutions between 1.7 Å and 2.1 Å in four different forms: (i) apo form, (ii) ternary complex with the nonhydrolyzable ATP analog AMPPNP and cofactor Ca2+, (iii) quaternary complex with Ca2+ and bound products AMP and phosphate, and (iv) binary product complex with AMP only. Analysis of the ATP (analog) binding mode explains the importance of several residues for activity and allows suggestion of a catalytic mechanism. The carboxylate group of E165 serves as a catalytic base and activates a water molecule, which is well positioned for nucleophilic attack on the terminal phosphate. Based on analysis of the two product complex structures in which AMP adopts different conformations, a substrate binding mode for ADP hydrolysis is proposed. This allows for an understanding of how the same hydrolytic site can be engaged in ATP and ADP but not AMP hydrolysis.
Keywords: ATP hydrolysis, CD39-like, ecto-enzyme, membrane protein, x-ray structure
Extracellular nucleotides like ATP function as signaling molecules that affect a wide range of physiological functions such as blood clotting, inflammation, immune reactions, pain perception, smooth muscle contraction, and cell proliferation (e.g., in embryonic development and cancer). Cellular responses are induced by binding to two classes of P2 receptors: G protein-coupled P2Y receptors and ligand-gated P2X ion channels (1–6). The “purinergic signaling” effects are converted and terminated via successive dephosphorylation of the nucleotides by a cascade of membrane-bound enzymes (7–11). Nucleoside triphosphate diphosphohydrolases (NTPDases, EC 3.6.1.5) and more specifically the cell surface-located NTPDase1–3 and -8 are the dominant ectonucleotidases relevant to P2 receptor-mediated signaling (10). They catalyze the sequential hydrolysis of the terminal γ- and β-phosphates from nucleoside triphosphates. The corresponding nucleoside monophosphate product can be further degraded to nucleoside and phosphate by ecto-5′-nucleotidase (5′NT or CD73) (11, 12). In addition to NTPs and NDPs also adenosine acts via specific receptors. As a result, the signaling effects of ATP are strongly influenced by the activities of ectonucleotidases depending on their differential expression patterns or catalytic properties.
NTPDase1–3 and -8 are glycoenzymes bound to the cytoplasmatic membrane by two helices close to the N and C termini (11). The interjacent catalytic domain belongs to the structural superfamily of actins, hsp70, and sugar kinases (13–15). It points toward the extracellular space and is stabilized by five invariant disulfide bridges (13, 16). Within the extracellular domain (ECD) five short sequence motifs of high conservation are noticeable among NTPDase family proteins (11). Of these apyrase conserved regions (ACR), ACR1 and ACR4 contain a prominent DXG motif and correspond to the β- and γ-phosphate binding loops of soluble members of the actin/hsp70/sugar kinase superfamily (17).
NTPDases differ significantly in product formation. This is of considerable relevance for the regulation of nucleotide signaling. NTPDase1, which corresponds to the lymphoid cell activation antigen CD39, hydrolyzes ATP processively to AMP [i.e., without release of the intermediate ADP from the active site (18, 19)]. The ectoenzyme is expressed by cells of the immune system, quiescent endothelial cells, vascular smooth muscle cells, and others. Because of its ability to hydrolyze both ATP and the platelet activator ADP to AMP, it blocks platelet aggregation and supports blood flow. Accordingly, NTPDase1-deficient knockout mice show disordered thromboregulation and increased infarct volumes upon experimental stroke (8, 20). NTPDase2, on the contrary, stands out for its preference for NTPs over NDPs. Upon hydrolysis of ATP, ADP would accumulate and only slowly be hydrolyzed to AMP. This behavior would potentially support platelet aggregation. Control of purinergic signaling by NTPDase2 has been implicated in the differentiation of stem cells in the nervous system (3, 21) and was recently shown to be responsible for initiation of embryonic eye development (5). Both the processivity of NTPDase1 and the high ATPase:ADPase ratio of NTPDase2 depend on native interactions of the transmembrane domains (19, 22–24). NTPDase3 and -8, which are expressed in brain and liver bile canaliculi, respectively, show intermediate patterns of product formation (10, 11). Compared with NTPDase1 and -2, the physiological relevance of these two enzymes is less well established.
Recently we described the bacterial production and kinetic characterization of soluble and active ECDs of NTPDase1–3 (24). This expression system has now led to the determination of the x-ray crystal structure of the ECD of Rattus norvegicus NTPDase2. This is the first structure of the Gda1/CD39 family of nucleoside phosphatases that currently includes >300 members. Based on the three complex structures with bound substrate analog, products, and cofactor as well as modeling of ADP into the active site, a possible catalytic mechanism is proposed, which allows for an understanding of the specificity of NTPDases for ATP and ADP but not AMP (11). Furthermore, the x-ray structure will allow for a rational design of NTPDase subtype-specific inhibitors for use in pharmacological research and as potential drugs for the treatment of thrombotic disorders, immune system diseases, and cancer (6, 25, 26).
Results
Overall Fold.
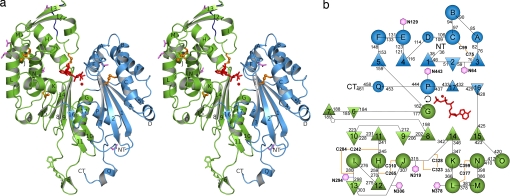
The protein that was subjected to crystallization trials consisted of residues T29 to S462 of R. norvegicus NTPDase2 and 23 additional N-terminal amino acids including a His6 tag as described (24). Residues P36 to Q288 and S296 to F461 are visible in the electron density. Residues R289–G295 most likely belong to a flexible loop. Proteolytical removal could be ruled out by SDS/PAGE analysis of crystals. However, the C-terminal amino acid S462 seems to be removed by proteolysis in all three structures because only a terminal carboxyl group could be modeled after F461. Between the two putative extracellular ends of the transmembrane helices and the termini of the ECD structure, only seven residues (N terminus) or one residue (C terminus) are missing. Hence, for the orientation given in Fig. 1a, the membrane would be located below the protein. The enzyme has a monomeric structure with dimensions of ≈75 Å × 60 Å × 45 Å (Fig. 1a). This is in agreement with the findings of us and others that cell-surface NTPDases are oligomeric in their membrane-bound form but monomeric when deglycosylated, detergent-solubilized from membranes, or expressed without transmembrane regions (19, 24, 27, 28).
Fig. 1.
Protein fold of the NTPDase2 ECD. (a) Cartoon diagram (Ca2+×AMPPNP complex). Domains I and II are colored in blue and green, respectively. A model conformation for the disordered loop (R289–S256) is indicated in dark blue. Disulfide bridges are shown in orange, Asn residues of possible N-glycosylation sites are in purple, and the cis peptide R191–P192 is in green. β-strands are numbered from 1 to 17. Helical segments are labeled A–Q. A, D, and Q are 310 helices. Helix Q is left-handed. Bound ligands are highlighted in red. (b) Topology diagram using the same coloring scheme as in a. β-Strands are represented by triangles, and helices are represented by circles. Possible N-glycosylation sites are shown as hexagons, and the cis proline is shown as a pentagon. The arrangement reflects the inner symmetry as revealed by superposition of the two domains (e.g., α-helices E and K can be superimposed whereas β-strand 10 does not have an equivalent). The two ligands mark the relative position of the active site between the two domains.
The ECD of NTPDase2 consists of two structural domains, each with an extended RNase H fold as expected from the relationship to the actin superfamily (Fig. 1b) (15). Both domains can be structurally aligned with an rmsd of 4.2 Å using 90 Cα atoms [program DALILITE (29)]. We classified the C-terminal helices of the two RNase H folds (i.e., G and P) to belong to the respective other structural domain based on the compactness of the resulting domains. Domain I (P36–S161 and K427–F461) is considerably smaller and more compact than domain II (G162–Q426). A DALI (29) search identified two members of the exopolyphosphatase/guanosine pentaphosphate phosphohydrolase (PPX/GPPA) family as closest structural homologs: 274 Cα atoms of the PPX enzyme from Escherichia coli (30) and the PPX/GPPA from Aquifex aeolicus (31) could be superimposed on NTPDase2 with rmsd values of 3.3 and 3.4 Å, respectively. Based on Psi-BLAST searches and fold recognition trials, the PPX/GPPA enzyme of A. aeolicus was previously identified as the closest structural homolog of NTPDase3 (13).
For the PPX/GPPA enzymes of A. aeolicus (31) and E. coli (30) open and closed forms have been analyzed, which differ by a domain rotation of up to 22.5° (32). The domain orientation in NTPDase2 most closely resembles the closed form of the E. coli enzyme. In NTPDase2 the β-sheet of the second RNase H motif is extended by the additional β-strand 10 antiparallel to β-strand 11 (Fig. 1). One short β-hairpin loop (β-strands 16 and 17) is inserted before the second interconnecting α-helix P. Helix Q is a left-handed 310 helix. In addition, the two-stranded antiparallel β-sheet made of β-strands 12 and 13 as well as the long extension that ends in a hairpin loop built from β-strands 6 and 7 do not have equivalents in the bacterial PPX/GPPA structures. This long extension is located close to the N and C termini and thus also close to the membrane. In this extension a cis peptide bond is formed between R191 and P192. Furthermore, it contains several exposed hydrophobic side chains (Y183, W185, V186, W189, and I190), also in the related cell-surface NTPDases [supporting information (SI) Fig. S1]. Possibly, this region interacts with or is inserted into the cell membrane and helps to anchor or orient the enzyme in its membrane-bound form. The positively charged side chains of K182, R188, and R191 could mediate interactions with the polar lipid head groups. A second explanation is that this structural element is necessary for oligomerization of the full-length protein.
The five disulfide pairings found in the crystal structure (1, C75–C99; 2, C242–C284; 3, C265–C310; 4, C323–C328; 5, C377–C399) (Fig. 1) agree with the ones determined previously (13, 16). All of the seven possible N-glycosylation sites of NTPDase2 have Asn residues exposed to the solvent, indicating that all are available for glycosylation. The crystal-packing interactions are unlikely to represent oligomerization interfaces of the full-length enzymes because either they do not form closed oligomer symmetries or the transmembrane helices would be located on different sides of the oligomer (data not shown). This is not unexpected because oligomerization of NTPDases is largely mediated by interactions of the transmembrane helices (11, 22, 23, 33).
Preparation of Ligand Complexes.
To allow identification of the substrate binding mode and elucidation of the catalytic mechanism, crystals were soaked with nonhydrolyzable adenine nucleotide analogs such as AMPCP, AMPPCP, ADPβS, ATPγS, and AMPPNP in combination with a variety of activating divalent metal ions [Mg2+, Ca2+, Zn2+, Sr2+, and Ba2+ (24)]. When the thio-substituted analogs were used, only the divalent metal ion, phosphate, and AMP, the final products of ATP or ADP hydrolysis, could be modeled into the electron density. Even with ADPβS, electron density corresponding to phosphate but not thiophosphate was found (data not shown). Using RP-HPLC (10, 24) it was shown that ATPγS and ADPβS are indeed weak substrates for NTPDase2 and degraded to AMP (data not shown). The hydrolysis of ATPγS or ADPβS in the crystal-soaking experiments could result from activity of the crystalline protein or from the activity of remnants of soluble NTPDase. An in-crystal activity test that is based on calcium phosphate deposition showed that the NTPDase2 ECD is active in the crystalline state (see Methods).
The generally more stable methylene-bridged substrate analogs AMPCP and AMPPCP as well as the imido-bridged ATP analog AMPPNP were not substrates in the RP-HPLC assay. In soaking or cocrystallization experiments with AMPCP or AMPPCP, no binding of the analogs or metal ions was observed. This indicates that the bridging oxygen atoms are necessary for binding of both substrates to the active site and that the metal ions do only bind to the enzyme in complex with the substrate. Only the isopolar ATP analog AMPPNP in complex with Ca2+ was able to bind to the active site without being hydrolyzed.
Active-Site Structure and Substrate Analog Binding.
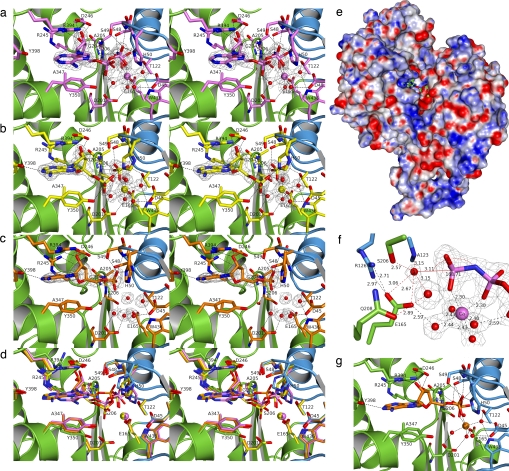
The binding site for the ATP analog and the divalent metal ion is located in the cleft between the two domains and well accessible from the surface (Figs. 1a and 2e). The adenine base of AMPPNP is sandwiched between Y350 and R394 (Fig. 2a). In the apo form R394 is only weakly defined (i.e., flexible), and also Y350 adopts a different conformation upon binding (Fig. 2d). R245 and A347 complete the hydrophobic binding pocket of the adenine base. In the Ca2+×AMPPNP complex no hydrogen bonds are formed between the adenine base and the protein. This binding mode, which largely depends on π-stacking interactions, explains the low base specificity of cell-surface NTPDases (14, 24).
Fig. 2.
Active-site structure. The cartoon fold is colored as in Fig. 1a. Omit electron density maps (Fo–Fc×ϕc) of the shown ligands and waters are contoured at 3.0σ. Hydrogen bonds involved in ligand binding are shown in black. (a) Ca2+×AMPPNP complex. (b) Ca2+×AMP×Pi complex. (c) AMP complex. (d) Superposition of the apo form (green/blue), the Ca2+×AMPPNP complex (purple), the Ca2+×AMP×Pi complex (yellow), and the AMP complex (orange). The cartoon fold is shown only for the apo form. (e) Molecular protein surface colored by electrostatic surface potential (Ca2+×AMPPNP) as calculated by PDB 2PQR software and the APBS plugin of PyMol. AMPPNP and Ca2+ are shown as sticks or balls. (f) Molecular environment of the terminal phosphate group of AMPPNP. The putative nucleophilic water is labeled “1,” and its hydrogen-bonding interactions with A123, E165, S206, and Q208 are drawn in red. R126 forms a salt bridge with E165. (g) Active site of a modeled productive Ca2+×ADP complex.
The ribose of ATP adopts the C2′ endo conformation. The 2′ hydroxyl group is hydrogen-bonded to the guanidino group of the base-stacking R394. R245 and D246 form hydrogen bonds with the 3′ hydroxyl group. The α-phosphate does not directly contact the protein. Several hydrogen bonds, however, are formed between the β- and γ-phosphate moieties and the backbone amides or side-chain functionalities of several residues belonging to the loops between strands 1 and 2 or 8 and 9: S48, S49, H50, G204, A205, and S206. In addition, the side-chain hydroxyl of T122 is involved in γ-phosphate binding. The β,γ-bridging imido group of AMPPNP is hydrogen-bonded to the backbone NH of G204 and the side-chain hydroxyl of S48, which acts as hydrogen bond acceptor. In the Ca2+×ATP binding mode, which probably closely resembles the Ca2+×AMPPNP binding mode, S48 is expected to function as a hydrogen bond donor. These two hydrogen bonds explain why the methylene-bridged ATP analog AMPPCP could not bind to the active site. Two nonbridging oxygens—one each from the β- and γ-phosphate groups—do not contact the protein. Instead they complete the coordination sphere of the Ca2+ cofactor, which is bound at the bottom of the cleft. The Ca2+ ion displays octahedral coordination by four water molecules and the phosphate oxygens and thus has no direct contacts to the protein. Rather, the four water molecules of the coordination sphere are positioned via interaction with D45, D201, T122, E165, and W436.
Product Binding Mode.
Because of the susceptibility of ADPβS toward enzymatic hydrolysis by the crystalline protein, the inability of AMPCP to bind to the active site, and the lack of commercial availability of AMPNP, no experimental ADP binding mode could yet be obtained. However, the products of the ADPase reaction are found in the active site of the quaternary Ca2+×AMP×Pi complex structure (Fig. 2b). The free orthophosphate occupies the same site as the terminal phosphate of the ATP analog and is still bound to the Ca2+ ion. This is also the site where a tungstate ion (used for phasing of the structure) bound to the enzyme (data not shown). AMP, the second product of ADP hydrolysis, adopts a conformation that differs significantly from the conformation of the AMP moiety of AMPPNP. Possibly as a result from electrostatic repulsion from the free phosphate, the α-phosphate is oriented more to the solvent. Hydrogen bonds are formed with two Ca2+-coordinating water molecules, the base-stacking Y350 and H50. Compared with the apo form and the Ca2+×AMPPNP structure H50 shows a flipped side-chain conformation (Fig. 2d). As a result of the altered α-phosphate binding, the hydrogen bonds from the ribose hydroxyl groups to R245 and R394 are weakened. Also, despite binding to the same hydrophobic base pocket as AMPPNP, the adenine base shows a different orientation, allowing it to form a single hydrogen bond with Y398.
The same adenine base binding mode is also observed in the binary AMP complex (Fig. 2c). In this complex the same hydrogen bonds from the ribose hydroxyl groups to R245, D246, and R394 are formed as in the Ca2+×AMPPNP complex. Without the electrostatic repulsion from the free orthophosphate the α-phosphate occupies a site that approximates to the β-phosphate binding site of AMPPNP (Fig. 2d). The same hydrogen bonds are formed with S48, S49, H50, and G204 (Fig. 2 a and c). Only the interaction with the divalent metal ion (which is not present in this structure) is missing.
Discussion
Catalytic Mechanism of ATP Hydrolysis.
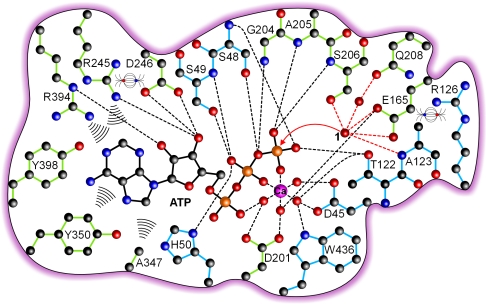
Elucidation of the binding mode for the isopolar ATP analog AMPPNP allows proposal of the catalytic mechanism in ATP hydrolysis. From the relation to the actin superfamily and studies on the related potato apyrase, hydrolysis is expected to occur by attack of a nucleophilic water on the substrate's terminal phosphate (34, 35). A water molecule that is positioned for direct in-line nucleophilic attack on the terminal phosphate is found with water 1 (Figs. 2f and 3). The position of water 1 is directed by interaction with the side-chain hydroxyl of S206, the backbone NH of A123, and the carboxyl group of E165. A fourth interaction is formed via water 2 with Q208. Water 1 is only 3.11 Å away from the phosphorus atom of the terminal phosphate. The angle formed by water 1, the phosphorus atom, and the β,γ-bridging nitrogen of AMPPNP is 168.7°. No other water or amino acid atom would be better positioned for a nucleophilic attack. The sole functionality that could act as a general base catalyst to activate water 1 is the carboxylate side chain of E165, 2.6 Å away. The side chain of E165 is positioned by forming a salt bridge to R126. The assignment of S206 and Q208 to be involved in positioning of the nucleophilic water, of E165 to function as catalytic base, and of R126 to be important for positioning of E165 is consistent with their absolute conservation among NTPDases and all other members of the PFAM Gda1/CD39 structural family. The importance of these residues for catalysis is also supported by mutagenesis studies (17, 36, 37): Even conservative substitution of the equivalents of E165 in NTPDase1 and -3 resulted in complete loss of activity. R143 in NTPDase3, which corresponds to R126, could be replaced by lysine but not by alanine. Substitution of the equivalents of S206 in NTPDase1 and -3 to alanine resulted in loss of activity. Furthermore, the residue corresponding to E165 was also identified as the catalytic base in the A. aeolicus PPX/GPPA enzyme after a product complex structure was solved at 2.7-Å resolution only recently (32).
Fig. 3.
Schematic active-site representation of the Michaelis complex for ATP hydrolysis and attack of the presumed nucleophile. The color scheme corresponds to Fig. 1. Hydrogen bonds are shown as dashed lines, hydrophobic interactions are shown as wave lines, and salt bridges are shown as electric field lines. The respective representation of a modeled ADP complex is shown in Fig. S2.
The divalent metal ion is thought to function as a catalyst by polarizing one of the P O bonds of the terminal phosphate. The importance of correct coordination of the cofactor is underpinned by the fact that mutation of residues involved in divalent metal ion binding (i.e., D45, D201, T122, E165, and W436) can result in loss of activity (15, 36–38) or alteration of substrate and/or cofactor specificity and affinity (38, 39). The negative charge of the transition state, which develops upon nucleophilic attack of the hydroxide ion corresponding to water 1, is stabilized through complexation of the electron-drawing divalent metal ion and by a series of hydrogen bonds from backbone NH groups, side-chain hydroxyls, and the H50 imidazole ring of the two phosphate binding loops.
O bonds of the terminal phosphate. The importance of correct coordination of the cofactor is underpinned by the fact that mutation of residues involved in divalent metal ion binding (i.e., D45, D201, T122, E165, and W436) can result in loss of activity (15, 36–38) or alteration of substrate and/or cofactor specificity and affinity (38, 39). The negative charge of the transition state, which develops upon nucleophilic attack of the hydroxide ion corresponding to water 1, is stabilized through complexation of the electron-drawing divalent metal ion and by a series of hydrogen bonds from backbone NH groups, side-chain hydroxyls, and the H50 imidazole ring of the two phosphate binding loops.
Modeling of a Productive ADP Binding Mode.
The finding that the Ca2+-bound phosphate of the Ca2+×AMP×Pi complex structure occupies the same site as AMPPNP's γ-phosphate suggests that the same hydrolytic site is used in ATP and ADP hydrolysis. To reach this hydrolytic site, the phosphate tail would have to adopt a more extended conformation than that observed in the ATP binding mode (Fig. 2a).
Examination of the binary AMP complex structure shows that this is possible: In a superposition of the nucleotide complex structures the α-phosphate of AMP is close to AMPPNP's β-phosphate (Fig. 1d). Thus, an ADP binding mode, which would allow usage of the same hydrolytic site as in ATP hydrolysis, is roughly outlined by AMP and the γ-phosphate of AMPPNP. Starting from the experimental coordinates a possible productive Ca2+×ADP binding mode was modeled (Fig. 2g and Fig. S2). In this model, the Ca2+ is coordinated by nonbridging oxygens of the α- and β-phosphates instead of β- and γ-phosphates of NTPs. As the β,γ-bridging imido group of the experimental Ca2+×AMPPNP structure, the α,β-bridging oxygen of the modeled Ca2+×ADP complex is making two hydrogen bonds to the protein (backbone NH and side-chain hydroxyl of S48). Thus, the model explains why, in addition to AMPPCP, also AMPCP could not bind to the active site.
In contrast to NTPs and NDPs, NMPs are not substrates for NTPDases because their phosphate groups cannot reach the hydrolytic site if the base is bound to the pocket between Y350 and R394 (Fig. 2c).
Implications for an Interdomain Motion.
The observation that the NTPDase2 ECD is active in the crystalline state argues against a large-domain motion being necessary for catalysis, because the crystal packing would not allow for large-domain reorientations. Apparently, the flexibility of the residues involved in substrate binding and catalysis (e.g., H50 in Fig. 2d) is sufficient for catalysis in the crystalline state. An interdomain motion has been discussed based on the relationship of NTPDases to actin and bacterial exopolyphosphatases and on the influence of the transmembrane helices on catalytic activity (13, 40).
Because several catalytic properties of NTPDases including the turnover rate, the ATPase:ADPase ratio, and the metal ion specificity change upon expression of the enzymes without the transmembrane helices (19, 23, 24, 28), it remains possible that an opening of the interdomain cleft is directed by the movement and interactions of the transmembrane regions and does occur only in the membrane-bound wild-type proteins. From Fig. 2 b and e it is conjecturable that the nucleotide product, Ca2+, and phosphate have to leave in a coordinated fashion from the active site of the NTPDase2 ECD. An opening of the active-site cleft may allow the presumably tightly bound phosphate product (Fig. 2b) to leave the active site more easily and independent of a preceding release of the nucleotide product, thereby enhancing the catalytic rate. Furthermore, a mechanism for phosphate release without loss of the nucleotide from the active site is of special importance for membrane-bound NTPDase1, where a processive hydrolysis from ATP to AMP occurs (18, 19).
Determination of the first structure of the NTPDase family in a substrate analog- and product-bound state allows a detailed assignment of functions to the individual ACR motifs. In addition, the structure gives several implications for structural and catalytic differences to related NTPDases and for the design of subtype-specific inhibitors as outlined in SI Discussion and Fig. S3.
In conclusion, with the ECD of the R. norvegicus NTPDase2 not only the first structure of cell-surface NTPDases, which constitute an essential component of extracellular purinergic signaling, but also of the largely membrane-bound PFAM Gda1/CD39 nucleoside phosphatase family (41) could be determined. The presented structure of NTPDase2 in an active state with bound cofactor, substrate analog, and products allows a deepened comprehension for the catalytic mechanism of signal conversion and inactivation in nucleotide-mediated cell signaling. Of special importance is the explanation of how the same hydrolytic mechanism is used in ATP and ADP hydrolysis. Experimental verification of the modeled ADP substrate binding mode will have to await successful structure solution of crystals soaked or cocrystallized with the imido-bridged ADP analog AMPNP or crystallization of inactive variants soaked with ADP.
Methods
Protein Purification and Crystallization.
Active NTPDase2 ECD protein was produced from bacterial inclusion bodies as described (24). The protein used for crystallization was stored at 4°C in 10 mM Tris·HCl, 1 mM NaN3, and 1 mM EDTA (pH 8.0) at a concentration of 2–3 mg/ml. Rod-like orthorhombic crystals (space group P212121, one molecule per asymmetric unit) were obtained at 4°C within 1 week to 1 month by hanging-drop vapor diffusion after mixing 1 μl of protein with 1 μl of reservoir solution containing 100 mM Hepes (pH 7.0–7.4) and 1–3% (wt/vol) PEG6000.
Derivatives were produced by soaking the crystals in a new drop with the composition of the original reservoir solution but with 5% (wt/vol) PEG6000 and the specific heavy atom salt or nucleotide. Crystals used for phasing were soaked overnight with 2 mM BaCl2 and 1.25 mM AMPCP (phasing native) or 4 mM Na2WO4 (tungstate derivative). Nucleotide complexes were produced by soaking the respective crystals for 1–3 h with 5 mM SrCl2 and 5 mM AMP (AMP complex), 20 mM CaCl2 and 11 mM ATPγS (Ca2+×AMP×Pi complex), or 10 mM CaCl2 and 10 mM AMPPNP (Ca2+×AMPPNP complex).
In-Crystal Activity Test.
To test whether the NTPDase2 ECD is active in the crystalline state, a protocol adopted from native PAGE activity staining (24) was developed. Crystals were extensively washed in reservoir solution with increasing amount of precipitant [from 2% to 20% (wt/vol) PEG6000] and 10 mM CaCl2. In the last step also 10 mM ATP was present in the crystal incubation solution. The hanging droplet with the crystal was then incubated at 4°C in a sealed crystal plate well. Around the crystals a slight white precipitate developed after 15 min that became very dense overnight. This precipitate is insoluble calcium phosphate, which is generated under these conditions from the nucleotidase activity of the crystals.
Data Collection and Structure Determination.
All x-ray data sets were collected at 100 K. Before cryocooling in liquid nitrogen crystals were transferred stepwise to a cryoprotection buffer. This was the reservoir solution or soaking solution plus 30% (vol/vol) PEG200 or 30% (vol/vol) glycerol. For structure determination, data sets were collected at beamlines ID13 and ID14.3 at the European Synchrotron Radiation Facility (Grenoble, France), BW7B and X12 at European Molecular Biology Laboratory/Deutsches Elektronen Synchrotron (Hamburg, Germany), and BL14.1 at Protein Structure Factory/Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung (Berlin, Germany). X-ray data sets were processed and scaled by using MOSFLM and SCALA (42).
The structure was determined by single isomorphous replacement with anomalous scattering using a single-site tungstate derivative. Initial models of the apo form, the binary AMP complex, the Ca2+× AMP×Pi complex, and the Ca2+×AMPPNP complex structure were refined to Rwork values of 17.5%, 17.0%, 16.2%, and 16.5% and Rfree values of 20.5%, 20.6%, 20.6%, and 21.0%, respectively. A detailed description of structure determination can be found in SI Text. The x-ray data, structure solution, and refinement statistics are listed in Table S1.
Modeling of a Productive Ca2+ × ADP Complex.
A model for the ADP-binding mode was built using the coordinates of AMP from the binary complex structure and of the phosphate and calcium ions from the quaternary complex. The ADP model was then copied into the structure of the quaternary complex, from which AMP, the phosphate ion, and two water molecules were deleted. During minimization of the whole system, the non-hydrogen protein atoms were tethered to their positions in the crystal structure. Futhermore, the stability of the ADP-binding mode was confirmed by a 100-ps molecular dynamics simulation without positional restraints. For all molecular modeling, calculations program MOE and the MMFF94x force field were used.
Supplementary Material
Acknowledgments.
We thank Prof. H. Zimmermann (University of Frankfurt, Frankfurt) for providing cDNA clones of rat NTPDases, and we thank the staff of beamlines ID13 and ID14.3 at the European Synchrotron Radiation Facility, BW7B and X12 at European Molecular Biology Laboratory/Deutsches Elektronen Synchrotron, and BL14.1 at Protein Structure Factory/Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung for beam time and support. The Deutsche Forschungsgemeinschaft is acknowledged for financial support.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3CJ1, 3CJ7, 3CJ9, and 3CJA).
This article contains supporting information online at www.pnas.org/cgi/content/full/0802535105/DCSupplemental.
References
- 1.Bours MJ, Swennen EL, Di Virgilio F, Cronstein BN, Dagnelie PC. Adenosine 5′-triphosphate and adenosine as endogenous signaling molecules in immunity and inflammation. Pharmacol Ther. 2006;112:358–404. doi: 10.1016/j.pharmthera.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 2.Burnstock G. Physiology and pathophysiology of purinergic neurotransmission. Physiol Rev. 2007;87:659–797. doi: 10.1152/physrev.00043.2006. [DOI] [PubMed] [Google Scholar]
- 3.Zimmermann H. Nucleotide signaling in nervous system development. Pflugers Arch Eur J Physiol. 2006;452:573–588. doi: 10.1007/s00424-006-0067-4. [DOI] [PubMed] [Google Scholar]
- 4.Stafford NP, Pink AE, White AE, Glenn JR, Heptinstall S. Mechanisms involved in adenosine triphosphate-induced platelet aggregation in whole blood. Arterioscler Thromb Vasc Biol. 2003;23:1928–1933. doi: 10.1161/01.ATV.0000089330.88461.D6. [DOI] [PubMed] [Google Scholar]
- 5.Massé K, Bhamra S, Eason R, Dale N, Jones EA. Purine-mediated signalling triggers eye development. Nature. 2007;449:1058–1062. doi: 10.1038/nature06189. [DOI] [PubMed] [Google Scholar]
- 6.Jackson SW, et al. Disordered purinergic signaling inhibits pathological angiogenesis in cd39/Entpd1-null mice. Am J Pathol. 2007;171:1395–1404. doi: 10.2353/ajpath.2007.070190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Todorov LD, et al. Neuronal release of soluble nucleotidases and their role in neurotransmitter inactivation. Nature. 1997;387:76–79. doi: 10.1038/387076a0. [DOI] [PubMed] [Google Scholar]
- 8.Enjyoji K, et al. Targeted disruption of cd39/ATP diphosphohydrolase results in disordered hemostasis and thromboregulation. Nat Med. 1999;5:1010–1017. doi: 10.1038/12447. [DOI] [PubMed] [Google Scholar]
- 9.Mizumoto N, et al. CD39 is the dominant Langerhans cell-associated ecto-NTPDase: Modulatory roles in inflammation and immune responsiveness. Nat Med. 2002;8:358–365. doi: 10.1038/nm0402-358. [DOI] [PubMed] [Google Scholar]
- 10.Kukulski F, et al. Comparative hydrolysis of P2 receptor agonists by NTPDases 1, 2, 3 and 8. Purinergic Signal. 2005;1:193–204. doi: 10.1007/s11302-005-6217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robson SC, Sévigny J, Zimmerman H. The E-NTPDase family of ectonucleotidases: Structure function relationships and pathophysiological significance. Purinergic Signal. 2006;2:409–430. doi: 10.1007/s11302-006-9003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knöfel T, Sträter N. X-ray structure of the Escherichia coli periplasmic 5′-nucleotidase containing a dimetal catalytic site. Nat Struct Biol. 1999;6:448–453. doi: 10.1038/8253. [DOI] [PubMed] [Google Scholar]
- 13.Ivanenkov VV, Meller J, Kirley TL. Characterization of disulfide bonds in human nucleoside triphosphate diphosphohydrolase 3 (NTPDase3): Implications for NTPDase structural modeling. Biochemistry. 2005;44:8998–9012. doi: 10.1021/bi047487z. [DOI] [PubMed] [Google Scholar]
- 14.Vorhoff T, Zimmermann H, Pelletier J, Sévigny J, Braun N. Cloning and characterization of the ecto-nucleotidase NTPDase3 from rat brain: Predicted secondary structure and relation to other members of the E-NTPDase family and actin. Purinergic Signal. 2005;1:259–270. doi: 10.1007/s11302-005-6314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith TM, Kirley TL. Site-directed mutagenesis of a human brain ecto-apyrase: Evidence that the E-type ATPases are related to the actin/heat shock 70/sugar kinase superfamily. Biochemistry. 1999;38:321–328. doi: 10.1021/bi9820457. [DOI] [PubMed] [Google Scholar]
- 16.Ivanenkov VV, Murphy-Piedmonte DM, Kirley TL. Bacterial expression, characterization, and disulfide bond determination of soluble human NTPDase6 (CD39L2) nucleotidase: Implications for structure and function. Biochemistry. 2003;42:11726–11735. doi: 10.1021/bi035137r. [DOI] [PubMed] [Google Scholar]
- 17.Kirley TL, Crawford PA, Smith TM. The structure of the nucleoside triphosphate diphosphohydrolases (NTPDases) as revealed by mutagenic and computational modeling analyses. Purinergic Signal. 2006;2:379–389. doi: 10.1007/s11302-005-5301-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heine P, Braun N, Heilbronn A, Zimmermann H. Functional characterization of rat ecto-ATPase and ecto-ATP diphosphohydrolase after heterologous expression in CHO cells. Eur J Biochem. 1999;262:102–107. doi: 10.1046/j.1432-1327.1999.00347.x. [DOI] [PubMed] [Google Scholar]
- 19.Chen W, Guidotti G. Soluble apyrases release ADP during ATP hydrolysis. Biochem Biophys Res Commun. 2001;282:90–95. doi: 10.1006/bbrc.2001.4555. [DOI] [PubMed] [Google Scholar]
- 20.Pinsky DJ, et al. Elucidation of the thromboregulatory role of CD39/ectoapyrase in the ischemic brain. J Clin Invest. 2002;109:1031–1040. doi: 10.1172/JCI10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mishra SK, et al. Extracellular nucleotide signaling in adult neural stem cells: Synergism with growth factor-mediated cellular proliferation. Development. 2006;133:675–684. doi: 10.1242/dev.02233. [DOI] [PubMed] [Google Scholar]
- 22.Grinthal A, Guidotti G. Transmembrane domains confer different substrate specificities and adenosine diphosphate hydrolysis mechanisms on CD39, CD39L1, and chimeras. Biochemistry. 2002;41:1947–1956. doi: 10.1021/bi015563h. [DOI] [PubMed] [Google Scholar]
- 23.Grinthal A, Guidotti G. CD39, NTPDase 1, is attached to the plasma membrane by two transmembrane domains. Why? Purinergic Signal. 2006;2:391–398. doi: 10.1007/s11302-005-5907-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zebisch M, Sträter N. Characterization of Rat NTPDase1, -2, and -3 ectodomains refolded from bacterial inclusion bodies. Biochemistry. 2007;46:11945–11956. doi: 10.1021/bi701103y. [DOI] [PubMed] [Google Scholar]
- 25.Gendron FP, et al. Purine signaling and potential new therapeutic approach: Possible outcomes of NTPDase inhibition. Curr Drug Targets. 2002;3:229–245. doi: 10.2174/1389450023347713. [DOI] [PubMed] [Google Scholar]
- 26.Müller CE, et al. Polyoxometalates—a new class of potent ecto-nucleoside triphosphate diphosphohydrolase (NTPDase) inhibitors. Bioorg Med Chem Lett. 2006;16:5943–5947. doi: 10.1016/j.bmcl.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Smith TM, Kirley TL. Glycosylation is essential for functional expression of a human brain ecto-apyrase. Biochemistry. 1999;38:1509–1516. doi: 10.1021/bi9821768. [DOI] [PubMed] [Google Scholar]
- 28.Knowles AF, Li C. Molecular cloning and characterization of expressed human ecto-nucleoside triphosphate diphosphohydrolase 8 (E-NTPDase 8) and its soluble extracellular domain. Biochemistry. 2006;45:7323–7333. doi: 10.1021/bi052268e. [DOI] [PubMed] [Google Scholar]
- 29.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 30.Rangarajan ES, et al. The structure of the exopolyphosphatase (PPX) from Escherichia coli O157:H7 suggests a binding mode for long polyphosphate chains. J Mol Biol. 2006;359:1249–1260. doi: 10.1016/j.jmb.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 31.Kristensen O, Laurberg M, Liljas A, Kastrup JS, Gajhede M. Structural characterization of the stringent response related exopolyphosphatase/guanosine pentaphosphate phosphohydrolase protein family. Biochemistry. 2004;43:8894–8900. doi: 10.1021/bi049083c. [DOI] [PubMed] [Google Scholar]
- 32.Kristensen O, Ross B, Gajhede M. Structure of the PPX/GPPA phosphatase from Aquifex aeolicus in complex with the alarmone ppGpp. J Mol Biol. 2008;375:1469–1476. doi: 10.1016/j.jmb.2007.11.073. [DOI] [PubMed] [Google Scholar]
- 33.Grinthal A, Guidotti G. Dynamic motions of CD39 transmembrane domains regulate and are regulated by the enzymatic active site. Biochemistry. 2004;43:13849–13858. doi: 10.1021/bi048644x. [DOI] [PubMed] [Google Scholar]
- 34.Flaherty KM, Wilbanks SM, Luca-Flaherty C, McKay DB. Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment. J Biol Chem. 1994;269:12899–12907. [PubMed] [Google Scholar]
- 35.Cohn M, Meek GA. The mechanism of hydrolysis of adenosine di- and tri-phosphate catalysed by potato apyrase. Biochem J. 1957;66:128–130. doi: 10.1042/bj0660128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang F, Hicks-Berger CA, Smith TM, Kirley TL. Site-directed mutagenesis of human nucleoside triphosphate diphosphohydrolase 3: The importance of residues in the apyrase conserved regions. Biochemistry. 2001;40:3943–3950. doi: 10.1021/bi002711f. [DOI] [PubMed] [Google Scholar]
- 37.Drosopoulos JH, et al. Site-directed mutagenesis of human endothelial cell ecto-ADPase/soluble CD39: Requirement of glutamate 174 and serine 218 for enzyme activity and inhibition of platelet recruitment. Biochemistry. 2000;39:6936–6943. doi: 10.1021/bi992581e. [DOI] [PubMed] [Google Scholar]
- 38.Drosopoulos JH. Roles of Asp54 and Asp213 in Ca2+ utilization by soluble human CD39/ecto-nucleotidase. Arch Biochem Biophys. 2002;406:85–95. doi: 10.1016/s0003-9861(02)00414-9. [DOI] [PubMed] [Google Scholar]
- 39.Smith TM, Lewis Carl SA, Kirley TL. Mutagenesis of two conserved tryptophan residues of the E-type ATPases: Inactivation and conversion of an ecto-apyrase to an ecto-NTPase. Biochemistry. 1999;38:5849–5857. doi: 10.1021/bi990171k. [DOI] [PubMed] [Google Scholar]
- 40.Musi E, Islam N, Drosopoulos JH. Constraints imposed by transmembrane domains affect enzymatic activity of membrane-associated human CD39/NTPDase1 mutants. Arch Biochem Biophys. 2007;461:30–39. doi: 10.1016/j.abb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 41.Bateman A, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.CCP4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.