Abstract
Mitotic chromosomes segregate at the ends of shortening spindle microtubules (MTs). In budding yeast, the Dam1 multiprotein complex supports this dynamic attachment, thereby contributing to accurate chromosome segregation. Purified Dam1 will track the end of a depolymerizing MT and can couple it to microbead transport in vitro. The processivity of such motions has been thought to depend on rings that the Dam1 complex can form around MTs, but the possibility that alternative coupling geometries contribute to these motilities has not been considered. Here, we demonstrate that both rings and nonencircling Dam1 oligomers can track MT ends and enable processive cargo movement in vitro. The coupling properties of these two assemblies are, however, quite different, so each may make a distinct contribution to chromosome motility.
Keywords: chromosome motions, kinetochore–microtubule interactions, microtubule end tracking
Mitotic chromosomes are attached to spindle fibers by a multiprotein complex, the kinetochore (1–3). Kinetochores bind strongly to the plus ends of spindle microtubules (MTs), but tubulin subunits can still be added or lost from these MT ends as the chromosomes move. Kinetochores are also sites where forces can be generated to move chromosomes either toward or away from the pole that a kinetochore faces, so this interface provides multiple important mitotic functions. Kinetochores include MT-dependent motors and several protein complexes that lack motor activity but can still bind to a MT wall or end. These nonmotor complexes too may be important for chromosome motion, because depolymerizing MTs can generate enough force to move chromosomes without the help of motors, both in vivo and in vitro (4–7).
The mechanism by which a kinetochore is coupled to the end of a shortening or lengthening MT is still unknown, but a 10-subunit protein complex from budding yeast kinetochores, called Dam1 or DASH, is important for chromosome–spindle attachments and spindle integrity in this organism (3). Purified Dam1 heterodecamers assemble around MTs into rings of 16–25 subunits (8, 9). At higher protein concentrations, both multiple rings and helices of Dam1 will form (10, 11).
In yeast spindles, Dam1 is found primarily at kinetochores, although some of it associates with spindle MTs (12). A careful estimate of the number of Dam1 subunits at metaphase kinetochores in budding yeast gave 16–20 heterodecamers (12), enough to form a single ring. The behavior of kinetochore-associated Dam1 fluorescence suggests that these subunits are stably attached to kinetochore MTs, showing little or no turnover. However, a second population of Dam1, which is dimly localized to spindle MTs, shows faster turnover, indicating the presence of a dynamic pool of complexes (12).
Oligomers of Dam1 can travel with a shortening MT in vitro (8), suggesting that this complex may be the primary MT-chromosome coupler in yeast. A bead coated with Dam1 will also associate with a MT in vitro and follow its end during either assembly or disassembly (13, 14), properties that are reminiscent of chromosome motions in vivo. Because Dam1 will assemble into rings that encircle MTs, all aspects of Dam1 behavior in vitro have so far been interpreted as a result of its forming ring-like structures.
To test this assumption and to gain insight into the structural requirements for Dam1 motility and coupling, we have examined MT depolymerization-dependent Dam1 behavior in vitro under conditions that are permissive for ring formation, or when ring formation is blocked or highly reduced. Our quantitative analyses demonstrate that not only rings but also other configurations of Dam1 can harness the energy available from MT depolymerization. These results strongly suggest that different oligomeric forms of Dam1 may contribute to mitotic chromosome motion.
Results
Dam1 Complexes That Track an MT End Move Steadily While Maintaining Their Size.
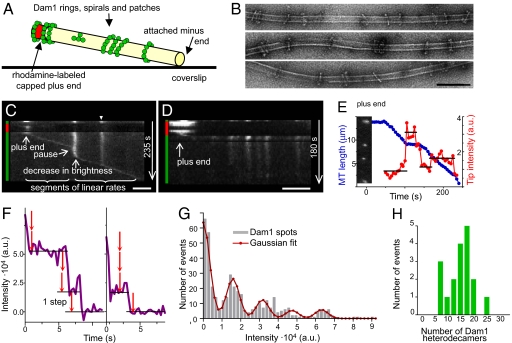
To study Dam1 motility at the ends of shortening MTs under conditions permissive for the assembly of ring-shaped oligomers, we used MTs that projected into solution but were firmly attached to a coverslip at their minus ends (Fig. 1A). MT plus ends were temporarily stabilized with caps of rhodaminated tubulin, assembled with a slowly hydrolyzable analog of GTP (GMPCPP), so depolymerization of the labile MT segment could be induced at will by dispersing its fluorescent cap [ref. 15; supporting information (SI) Text, Part 1]. Dam1 labeled with Alexa488 formed static green dots with various intensities along these stabilized MTs. Previous studies suggest that such dots could correspond to single rings, stacks of rings, or short spirals (11). To determine the structure of the Dam1 oligomers in our experimental system, we examined samples from these chambers by EM, but the MTs that projected into solution tended to break, so only the shortest MTs, perhaps those that were GMPCPP-stabilized, survived on EM grids. Furthermore, at the range of Dam1 concentrations where there were only a few fluorescent dots on MTs 10–20 μm long (2–20 nM), it was impossible to find representative samples of Dam1–MT complexes by EM, where the field of view is small. We therefore mixed taxol-stabilized MTs with Dam1 at higher Dam1 concentrations (200–350 nM). The resulting MTs were uniformly green in the fluorescence microscope, and in the EM, both single and double rings were now readily seen on the MTs (Fig. 1B), suggesting that these are the most common structures that form under our experimental conditions.
Fig. 1.
Quantitative analysis of the tracking Dam1 complex. (A) Schematic of the experimental system (not to scale). (B) EM of Dam1 oligomers that assembled on taxol-stabilized MTs. (Scale bar, 100 nm.) (C) Kymograph of Alexa488-Dam1 on a depolymerizing MT in motility buffer with βME. The majority of dots formed by wild-type Dam1 do not move until a MT end comes by (example marked with arrowhead; diffusion <10−13 cm2/sec). Colored vertical bars on these and other kymographs indicate the fluorescent channel with which images were acquired. (Scale bar, 2 μm.) (D) Without βME, Dam1 does not track the shortening MT end and inhibits MT depolymerization. (Scale bar, 2 μm.) (E) Kinetics of a distal Dam1 dot and its normalized intensity (arbitrary units) for the same MT as in C. Inset shows the initial image of this MT decorated with Alexa488-Dam1. Horizontal lines show the average intensity for a given time interval. Variations of the intensities within these intervals are not statistically significant, because not all captured images of the jiggling MTs are in perfect focus. (F) Typical photobleaching curves for coverglass attached, dim Dam1 dots. The step size in these images corresponds to a value determined by two different step-identifying algorithms. (G) Intensity histogram for Dam1 dots (average of 17 experimental curves with total of 486 data points) and its Gaussian fitting. (H) The intensities of tracking complexes show that they have a preferred size.
When the stabilizing cap was dispersed on our segmented MTs decorated with Alexa488-Dam1, a green dot followed the shortening MT end (Movie S1, Fig. 1C), as described in a different system (8). This tracking of the MT end by Dam1 complexes depended on our addition of 2-mercaptoethanol (βME). Without this reducing agent, Alexa488-Dam1 still formed dots on MTs, but MT depolymerization was severely inhibited, and the dots did not move, indicating an importance for sulfhydryl (SH) groups in MT-mediated Dam1 sliding (Fig. 1D). In the presence of βME, a distal Dam1 spot tracked the shortening MT end with a rather constant speed (Fig. 1E, and Fig. S1). The brightness of a tip-associated Dam1 complex was also relatively constant during the segments of linear motions on MTs free of other complexes, indicating the maintenance of a constant end-tracking structure. However, it was also apparent that steady Dam1 tracking was sometimes interrupted; pauses usually occurred when the tracking complex encountered a bright lattice-bound complex. Before the resumption of tracking, spot brightness usually returned to its original level (Fig. 1 C and E and Fig. S1), strongly suggesting that the tracking complex had a preferred size. These observations prompted us to examine the size of the tracking complexes and the nature of their pausing.
An End-Tracking Dam1 Complex Can Be a Single Ring but Not a Larger Complex.
To assess the number of Dam1 subunits in the tracking complexes, we first quantified the fluorescence of motionless coverslip-associated dots of Alexa488-Dam1 and measured the kinetics of their photobleaching. When dim dots were photobleached with the 488-nm line from a stable Argon-ion laser, their intensity decreased in visible steps (Fig. 1F). To test this assertion and to identify the size of a single step, we used two algorithms: Gaussian fitting of the fluorescence-intensity histogram (16) and the pairwise distance difference (PDF) method (17) (SI Text, Part 2). Both methods revealed the presence of statistically significant stepwise changes in fluorescent intensity (P < 10−5). The unit brightness of Alexa488 fluorophores in our system was 15,800 ± 1,200 and 16,362 ± 1,990 arbitrary units, as determined with these two algorithms, respectively (Fig. 1G). The molar ratio of fluorophores to protein for our labeled Dam1 was 2.1 ± 0.1, so spot brightness before bleaching provided a measure of the number of Dam1 subunits in that oligomer. For example, the dim, motionless spots attached to a coverslips contained 1.9 ± 0.4 (n = 44) heterodecamers, suggesting a dimer. In the same experimental chambers and under identical conditions, the distribution of intensities of the spots that tracked with shortening MTs showed a significant grouping (Fig. 1H): they contained 19 ± 3 (n = 18) heterodecamers. The largest moving complex observed contained ≈25 subunits. We concluded that the oligomers that tracked shortening MT ends had a composition consistent with a single but not a double ring.
A Moving Dam1 Complex Can Shed Subunits When It Encounters a Large Static Complex.
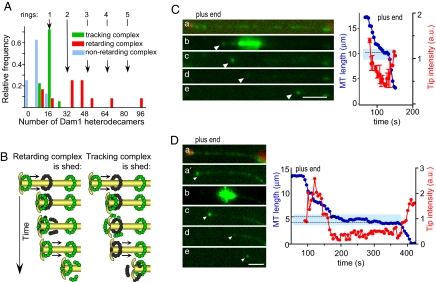
Although the tracking Dam1 complex moved with a constant speed on a MT wall that was free of other complexes, its behavior was more complex when it encountered another Dam1 oligomer. Occasionally, the MT wall-associated Dam1 dots were “collected” by the shortening end, as in ref. 8, but such dots were usually dimmer than the tip complex and contained on average 9 ± 1 Dam1 heterodecamers (Fig. 2A). These dots did not slow MT shortening significantly: the ratio of average rates for segments with and without such dots was 1.2 ± 0.1 (n = 94 and 85, respectively). For examples, see the dimmer dots on Fig. 1 C and E and Fig. S1A. However, Dam1 complexes with intensities similar to or brighter than that of the tracking complex usually halted MT depolymerization, although it resumed later with a highly similar speed (e.g., brighter dots on Fig. 1 C and E and Fig. S1). In 90% of such cases (n = 29), the resumption of motion occurred only after a decrease in Alexa488-Dam1 brightness, indicating that large complexes could not track MT ends processively. Together, these quantitative analyses strongly suggest that when a tracking ring encounters one or more static MT wall-associated rings (Fig. 2A), a shedding of excess Dam1 is required for tracking to resume.
Fig. 2.
Shedding of Dam1 complexes under the depolymerization force. (A) Intensity histogram of tracking (in green, same data as in Fig. 1H) and MT wall-associated complexes that retard MT depolymerization (in red) or are collected without a significant change in the tracking rate (blue). (B) Two models to explain Dam1 shedding. Bleached rings are black. (C and D) Photobleaching of MT-associated Alexa488-Dam1 complexes (MT plus end cap is red, Alexa488-Dam1 is green). Target area (blue box in graphs) was illuminated with a 488-nm laser (b in C and D). Depolymerization paused in the bleached area, as is common in unbleached areas with dots, suggesting that photobleaching did not change these properties of the complexes. (Scale bars, 3 μm.) Arrowheads point to tip-associated complexes.
We have used laser photobleaching to ask whether the tracking Dam1 complexes displaced “downstream” wall-associated ones or vice versa. If the moving (upstream) complex displaced the motionless downstream one, then the brightness of the tracking signal should be constant and unaffected by the intensity of the downstream complex (Fig. 2B Left), whereas if the downstream complex displaces upstream, the opposite should prevail (Fig. 2B Right). To discriminate between these possibilities, we photobleached MT-associated Alexa488–Dam1 complexes that were not at the MT tip. Photobleached static complexes on MT walls remained dim for minutes, confirming a slow rate of exchange between soluble and MT-bound Dam1 (11). After such photobleaching, we induced tubulin depolymerization. The brightness of the tip-associated signal decreased as it moved into a bleached area, and fluorescence reappeared only after MT depolymerization continued into an unbleached region and the tracking complex encountered a bright, static complex (Fig. 2 C and D).
When Dam1 Is in Solution, a Tip-Associated Microbead Slides Along the MT Surface.
We then analyzed the cargo-moving properties of Dam1 complexes. Previous studies have demonstrated that microbeads coated with Dam1 protein can move processively at the ends of shortening MTs (8, 13, 14). In our system, too, Dam1-coated beads bound readily to MTs preincubated with Alexa488-Dam1, and when MTs were induced to disassemble, the beads moved processively (Movie S2).
Two observations suggested that the beads in our system tracked the MT end while stably attached to a Dam1 ring. First, at reduced levels of Dam1 decoration, the beads moved processively, reminiscent of the steady tracking by tip-associated complexes on MT segments free of Dam1 decoration. However, with higher Dam1 density on the MTs, the tracking beads sometimes encountered a static Dam1 complex. Such encounters frequently resulted in bead detachment (Fig. S2A), as expected from the results on the shedding of a tracking Dam1 complex described above. Second, we tested the ring-attachment hypothesis by examining the 3D motions of beads that were tracking a shortening MT end. If the beads were bound to MTs by rings, these motions should comprise a simple sliding along the MT wall, so any point on the bead's surface, not just its center, should move parallel to the MT axis. To test this prediction, we spiked one side of our beads with fluorescently labeled Dam1, then saturated the remaining binding sites with unlabeled Dam1. This process produced beads with a uniform distribution of Dam1 but unevenly distributed fluorescence. When these Dam1-coated beads were allowed to associate with MTs in the presence of soluble Dam1, they moved with simple sliding motions, as expected, if these beads were being pulled by rings (Movie S3).
Dam1-Coated Beads Can Track MT Ends in the Absence of Ring Formation.
We have asked whether rings are required for bead transport on shortening MTs by coating microbeads with Alexa488-Dam1, then thoroughly rinsing them free of soluble protein (SI Text, Part 1). These beads attached only to the GMPCPP portions of our segmented MTs, not to their GDP-containing segments (Fig. S2B). Even when the beads were held against the MT wall with the laser trap, without soluble Dam1, they failed to form a firm association with the GDP-MT wall. This suggested that without Dam1 in solution, the bead-MT links were formed by only a few bead-associated Dam1 heterodecamers, rather than by a multisubunit encircling ring. Such weak binding was apparently stabilized by the higher affinity of Dam1 for a GMPCPP-tubulin-containing lattice at the MT caps (11).
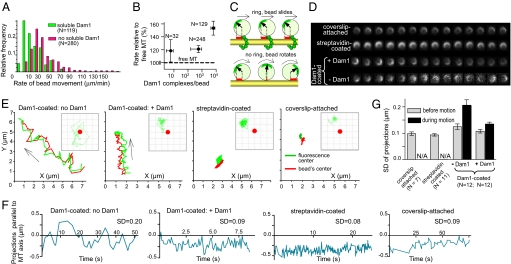
When the GMPCPP caps were photodispersed, 46% of 420 beads fell off, but the rest moved steadily toward the MT minus ends at 29.9 ± 1.2 μm/min (Movie S4). Such MT end tracking by Dam1-coated beads in the absence of soluble Dam1 has been interpreted previously as a result of bead-associated rings, which somehow formed around the MTs from the bead-bound Dam1 (13, 14). If this interpretation were correct, then adding soluble Dam1 to this system should have no effect on the system's kinetics, but in the presence of soluble Dam1, the speed of bead movement dropped to 7.9 ± 1.0 μm/min (Fig. 3A, Fig. S2C).
Fig. 3.
Dam1-mediated motions of microbeads. (A) A histogram of rates for MT disassembly-driven movements of Dam1-coated beads. When Dam1 is in solution, beads move significantly slower than when soluble Dam1 is absent. (B) The relative rates of MT depolymerization-dependent movement of Dam1-coated beads in the absence of soluble Dam1 for different average numbers of Dam1 molecules bound to each bead, plotted on a semilog scale. (C) Schematic of the experimental system (not to scale) to examine beads rotational mobility with (Upper) and without (Lower) ring-shaped coupling. Red marks represent projections on the MT axis of the vector from the bead's center to the brightest fluorescent dot. These projections were used to compare the rotational freedom of different beads, as shown in E–G. The rotational mobilities of 1 μm streptavidin and Dam1-coated beads attached to MTs were monitored before and after the initiation of MT depolymerization (only Dam1-coated beads move). Dam1-coated beads were examined with or without soluble Dam1; the streptavidin-coated beads were attached to biotinylated MTs; beads that stuck to the coverslip were used as an additional control. Motions of one bead from each of these categories are shown in D (time-lapse images) and E [trajectories of the center of the bead (red) and the brightest fluorescent dot on its surface (green curves) in the imaging XY plane relative to the reference point (0, 0)]. Insets in E show movement of the brightest fluorescent dot (green tracks) relative to the bead's center (red circle). Grid size 0.25 micron. (F) The graphs show the projections on the MT axis of the vector that connects the bead's center and the brightest fluorescent dot on the bead's surface for these four beads. SD for such curves were used as a measure of the bead's rotational activity, summarized in G.
Remarkably, without soluble Dam1, the Dam1-coated beads increased the rate of MT depolymerization, which in our system was 21.9 ± 2.0 μm/min, n = 57. When more Dam1 was bound to the beads, their average speed increased even further (Fig. 3B), contrary to expectations based on the hypothesis that rings can form from bead-associated Dam1. Furthermore, beads coated with the S4D phosphomimetic Dam1 complex, in which the oligomerization of Dam1 heterodecamers is reduced (18), also tracked shortening MT ends (Fig. S2D). Finally, MT end-tracking by Dam1-associated beads occurred even in the absence of βME (Fig. S2D), indicating a biochemically different requirement for this type of motility than when rings can form. Together, these results strongly suggest that Dam1-coated beads can be transported by depolymerizing MTs in the absence of ring assembly. Previous studies using such beads in the absence of soluble Dam1 and βME (13, 14) are likely to be pertinent to this ring-independent motility (SI Text, Part 3).
Dam1-Coated Beads Roll Rather Than Slide When Soluble Dam1 Is Absent.
Previous reports have shown that proteins other than Dam1 can transduce the energy from MT depolymerization into bead motility (19). A ring-like structure was most unlikely to have contributed to the nucleotide-independent motions of these beads, which were coated with dynein, kinesin, or the chimeric kinesin NK350 (19). It has been suggested that this kind of bead motility resulted from a bead's rolling in front of the protofilaments that bend as the MT shortened (20, 21). To test this model, we used the above-described approach with unevenly fluorescent beads. In the absence of soluble Dam1, some of the Dam1-coated beads obviously rotated while moving with a shortening MT (Fig. 3 C and D, Movie S5), a behavior that is incompatible with ring-based attachment. The rolling is obvious, however, only when the plane of bead rotation remains perpendicular to the optical axis. Because the beads oriented randomly, and thermal motions were significant, we quantified the degree of bead rotation by determining the projections of their brightest spot onto the MT axis in the image plane (Fig. 3 C and E–G). Beads moving with depolymerizing MTs showed significantly higher rotational mobility than the same beads before their start of directed motion, beads monitored in the presence of soluble Dam1, beads that were statically attached to MTs via biotin-streptavidin link, or than beads that were stuck to the coverglass (Fig. 3G). Because mitotic chromosomes do not rotate while moving, it appears that the mechanism by which Dam1 supports bead motion in the absence of rings is unlikely to be directly relevant to the situation in vivo.
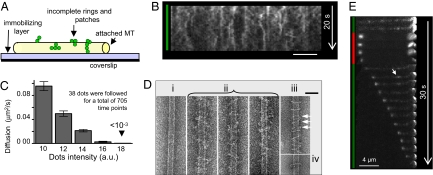
Nonencircling Dam1 Assemblies Have Diverse Shapes and Diffuse Rapidly.
The above results demonstrated that Dam1 heterodecamers can bind MTs and even support bead transport in the absence of ring formation. Structural studies, however, have suggested the ring formation is highly favored, because nonring complexes are infrequently seen by EM [Fig. 1B (10, 11)]. There may, however, be a bias toward the visualization of rings if they preferentially survive specimen preparation or are easier to see. To ask whether Dam1 will bind MTs when ring formation is prevented, we attached biotinylated MTs to a streptavidin-coated coverslip, thereby blocking the formation of MT-encircling structures (Fig. 4A). Alexa488-Dam1 protein still bound to these MTs, forming fluorescent dots that diffused rapidly, frequently merging and separating (Fig. 4B). As with ring sliding, these motions depended on the presence of βME (Fig. S3). The rate of spot movement decreased with increasing fluorescence intensity, as expected for randomly diffusing particles with different numbers of bonds to their substratum (Fig. 4C). EM of these preparations, which were lightly fixed to stabilize weak interactions, revealed complexes with various sizes and shapes, some of which resembled arcs (Fig. 4D). Thus, Dam1 heterodecamers can bind MTs not only as encircling assemblies (10, 11) but also as mobile, structurally diverse complexes, which we call “patches.”
Fig. 4.
Motility of MT-associated Dam1 when ring formation is prevented or reduced. (A) Schematic of the experimental system to sterically prevent the formation of rings (not to scale). (B) A kymograph, which illustrates changes over time in the positions and intensities of Alexa488-Dam1 complexes on a coverglass-attached MT. (Scale bar, 4 μm.) (C) Diffusion coefficients of Dam1 patches plotted against their initial intensities. (D) EM images of negatively stained MTs attached to an immobilizing layer in the absence (i) or presence of Dam1 (ii, iii). Image in iii was obtained from MTs first incubated with soluble Dam1 and then attached to the immobilizing layer in Dam1's presence, so both rings and nonencircling complexes (arrows) could assemble. Example of a Dam1 ring formed on MTs suspended in Dam1 solution is shown in iv. (Scale bar, 50 nm.) (E) Time lapse of a depolymerizing MT coated with Alexa488-labeled S4D-Dam1. Unlike with wild-type Dam1, the MT-wall associated dots of mutant protein are faint and diffusive (arrow), but they still show increased brightness at the shortening end.
Nonencircling Dam1 Assemblies Can Follow a Depolymerizing MT End.
To examine the structural requirements for MT end-tracking by Dam1 in the absence of beads, we analyzed the properties of a Dam1 mutant in which ring formation is reduced (18). Alexa488-labeled S4D mutant Dam1 bound readily to MTs that projected into solution, consistent with its reported affinity for these polymers (11). It failed, however, to form the discrete, well contrasted MT-associated dots that were characteristic of the wild-type complex. Mutant protein decorated MTs more evenly, and the areas of increased brightness appeared to diffuse more rapidly than wild-type Dam1 complexes. When MTs coated with this mutant Dam1 depolymerized, the distal Dam1 dot moved processively with the shortening MT ends at 25.1 ± 2.9 μm/min, n = 38 (Fig. 4E). Furthermore, with this mutant, we observed some events similar to those originally described for wild-type Dam1 (8, 11), such as MT bundling and motions indicative of force-coupling properties. For example, a shortening MT end attached to the wall of another MT by a S4D Dam1 dot exerted a pulling force, so the attached MT bowed (Movie S6). Clearly, visible manifestations of MT end tracking and force generation by Dam1 protein complexes do not require the efficient formation of rings.
Discussion
Kinetic and Coupling Properties of the Dam1 Oligomers in Vitro.
Previous work has demonstrated different MT-dependent motions of the Dam1 complexes in vitro (8, 13, 14). Although EM studies imply that a MT encircling is the prevalent form of Dam1 oligomerization [Fig. 1B (8, 10)], it appears that a noticeable fraction of Dam1 complexes formed in vitro consists of incomplete rings and patches. Our discovery that these small complexes can exhibit all known types of Dam1-mediated motility, most notably the tracking of MT ends and microbead transport, highlights the importance of detailed examination of all aspects of these phenomena, so that each of these different oligomeric forms receives an accurate description.
Our evidence that the majority of complexes that track shortening MT ends in vitro are single rings is not compelling, but it is strong and similar in nature to the arguments that suggest the presence of rings at yeast kinetochores (12). Most importantly, the tracking complex has a number of subunits similar to the ring size seen by EM. The tracking complex is also remarkably robust: it travels with a constant speed and unchanged brightness over MT segments free of other complexes, and it can even collect smaller Dam1 complexes without significant slowing. A microbead carried by the tracking complex shows no significant rotation, consistent with a sliding motion, which a ring must exhibit for processive motility. There are no detectable saltatory motions of the complexes that are large enough to form a ring, both when they track the MT end and when they are bound to the MT wall (Fig. 1C and Fig. S1A), consistent with a notion that these complexes adhere strongly to the MTs (11, 22).
A previous study of Dam1 bound to GMPCPP MTs that were depolymerized by the destabilizing kinesin XMCAK1 found a higher density of rings at the MT ends than on the walls; this was thought to result from the accumulation of sliding rings that had stacked together (11). Strongly bound rings, however, should not collect easily, consistent with our finding that the tracking complexes did not contain enough subunits to form more than one ring (Fig. 1H). Indeed, in this work, where GDP-MTs were induced to depolymerize by removal of a stable cap, the tracking Dam1 complex disintegrated when it encountered another such complex (Fig. 2). We hypothesize that the increased density of rings at the ends of shortening MTs, reported in ref. 11 reflected the retardation of MT depolymerization by large Dam1 stacks rather then their collection. The Dam1 shedding that occurs during pausing might be detrimental to processive cargo movement (Fig. S2A), but because the intracellular Dam1 protein level is low (10, 12), such events are unlikely to occur during kinetochore motions in vivo.
Unlike ring-size Dam1 complexes, Dam1 patches diffuse fast. They too can travel with the shortening MT ends (Figs. 1H and 4E). The dimly labeled population of Dam1 complexes along spindle MTs (12) and the rapidly diffusing complexes seen along kinetochore MTs (6) are likely to represent these nonring Dam1 complexes in vivo. Future work should focus on determining detailed kinetics properties of Dam1 patches, such as the rate at which they track MT ends and the degree of their collection, because these characteristics are vital for determining whether they could serve as good kinetochore couplers. Although the Dam1-coated beads can track shortening MT ends in the absence of Dam1 rings, the beads roll on the MT surface rather than slide. We take this as an indication that the Dam1 patches cannot ensure the kind of coupling required for the translational motions of chromosomes, but further work is required to address this important issue.
On the Nature of Dam1 Complexes That Function at the Budding Yeast Kinetochore.
If Dam1 nonencircling complexes can support MT-dependent motility in vitro, as our work suggests, could they carry on this task in live cells? The deletion of any component of the Dam1 complex is lethal in budding yeasts, but cells with the S4D mutation are viable (3). Such cells do, however, have defective kinetochore-MT attachments, as evidenced by lagging chromosomes in anaphase (23). Based on this phenotype and our own observations, we suggest that these ring-deficient Dam1 complexes can support depolymerization-dependent cargo motions, but they are less effective than rings in some aspect of a task that is important for accurate chromosome segregation.
Early in mitosis, the two forms of Dam1 oligomers may coexist dynamically to facilitate the formation of rings on properly bioriented kinetochores, during which time the ringless geometries of Dam1 may provide auxiliary functions for chromosome motilities. However, the ring-forming Dam1-coupler may be preferred for maintaining a force-transducing link, because these attachments have to be very stable (24). Interestingly, the number of Dam1 complexes per kinetochore in fission yeast is estimated to be less than that required for a full ring, suggesting that nonencircling assemblies may be the preferred functional form in these cells (25); other MT end-tracking proteins may also be involved (26). In mammalian cells, where a close Dam1 homolog has not been found, processive kinetochore-MT attachment may be facilitated by fibrillar complexes rather than rings (J.R.M., unpublished work). Future studies should focus on defining the biophysical properties of these structurally different couplers, so that we may learn about their respective strengths and limitations.
Materials and Methods
Reagents and Experimental Conditions.
Bacterially expressed Dam1 complexes were purified and labeled as in refs. 10 and 11. After dialysis and centrifugation (130,000 × g, 15 min), Dam1 was kept on ice; immediately before experiment, it was diluted to 1–20 nM into “motility” buffer: 80 mM K-Pipes, pH 6.9; 1 mM EGTA; 4 mM MgCl2; 1–2 mM DTT; casein (0.5 mg/ml); BSA (4 mg/ml); and 0.5–1% βME. All in vitro assays were performed as in ref. 15, unless stated otherwise. To sterically block ring formation, MTs were grown from a mixture of unlabeled and biotinylated tubulin (2.5:1), stabilized with taxol, and attached to a streptavidin-containing coat on a coverglass (0.5 mg/ml biotinylated BSA followed by rinsing, then by 0.5 mg/ml streptavidin). In all other experiments, Chlamydomonas axonemes or Tetrahymena pellicles were affixed to a coverslip to nucleate MT growth with 1 mM GTP. MTs were subsequently stabilized by capping them with rhodamine-labeled tubulin in 0.5 mM GMPCPP (15). Disassembly of such capped MTs was induced with the light from an HBO100 lamp, filtered for Texas red. Under conditions in which the minus MT end was also dynamic, the movement of end-proximal Dam1 dots occurred similarly at both MT ends. We used 0.52-μm silica beads (carboxylate-modified; Bangs Laboratories) and polystyrene beads with covalently bound streptavidin (0.44 μm from Spherotech and 0.51 μm from Bangs Laboratories). The results from these different beads were not significantly different, so they were grouped. Detailed information about other experimental protocols, preparation of the Dam1-coated beads, data acquisition, and EM is provided in SI Text, Part 1, and Fig. S4.
Supplementary Material
Acknowledgments.
We thank N. Gudimchuk for analysis of photobleaching steps, M. Porter (University of Minnesota, Minneapolis) for the kind gift of Chlamydomonas axonemes, and A. I. Vorobjev for support. EM was done in the University of Colorado, Boulder, laboratory for 3D microscopy, with M. Morphew's help. This work was supported by National Institutes of Health Grants GM033787 and RR000592, to J.R.M., who was a Research Professor of the American Cancer Society; by U.S. Civilian Research and Development Foundation Grant CGP2006B#2863 (to F.I.A. and J.R.M.); and by Russian Academy of Sciences Grant MCB RAS.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801811105/DCSupplemental.
References
- 1.McIntosh JR, Grishchuk EL, West RR. Chromosome-microtubule interactions during mitosis. Annu Rev Cell Dev Biol. 2002;18:193–219. doi: 10.1146/annurev.cellbio.18.032002.132412. [DOI] [PubMed] [Google Scholar]
- 2.Maiato H, DeLuca J, Salmon ED, Earnshaw WC. The dynamic kinetochore-microtubule interface. J Cell Sci. 2004;117:5461–5477. doi: 10.1242/jcs.01536. [DOI] [PubMed] [Google Scholar]
- 3.Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu Rev Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- 4.Coue M, Lombillo VA, McIntosh JR. Microtubule depolymerization promotes particle and chromosome movement in vitro. J Cell Biol. 1991;112:1165–1175. doi: 10.1083/jcb.112.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grishchuk EL, McIntosh JR. Microtubule depolymerization can drive poleward chromosome motion in fission yeast. EMBO J. 2006;25:4888–4896. doi: 10.1038/sj.emboj.7601353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka K, Kitamura E, Kitamura Y, Tanaka TU. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J Cell Biol. 2007;178:269–281. doi: 10.1083/jcb.200702141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franco A, Meadows JC, Millar JB. The Dam1/DASH complex is required for the retrieval of unclustered kinetochores in fission yeast. J Cell Sci. 2007;120:3345–3351. doi: 10.1242/jcs.013698. [DOI] [PubMed] [Google Scholar]
- 8.Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–569. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- 9.Miranda JJ, King DS, Harrison SC. Protein arms in the kinetochore-microtubule interface of the yeast DASH complex. Mol Biol Cell. 2007;18:2503–2510. doi: 10.1091/mbc.E07-02-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miranda JJ, De WP, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- 11.Westermann S, et al. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 12.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci USA. 2006;103:9873–9878. doi: 10.1073/pnas.0602249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franck AD, et al. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat Cell Biol. 2007;9:832–837. doi: 10.1038/ncb1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR. Force production by disassembling microtubules. Nature. 2005;438:384–388. doi: 10.1038/nature04132. [DOI] [PubMed] [Google Scholar]
- 16.Park M, Kim H-H, Kim D, Song NW. Counting the number of fluorophores labeled in biomolecules by observing the fluorescence-intensity transient of a single molecule. Bull Chem Soc Jpn. 2005;78:1612–1618. [Google Scholar]
- 17.Block SM, Svoboda K. Analysis of high resolution recordings of motor movement. Biophys J. 1995;68:230S–239S. [PMC free article] [PubMed] [Google Scholar]
- 18.Wang HW, et al. Architecture of the Dam1 kinetochore ring complex and implications for microtubule-driven assembly and force-coupling mechanisms. Nat Struct Mol. Biol. 2007;14:721–726. doi: 10.1038/nsmb1274. [DOI] [PubMed] [Google Scholar]
- 19.Lombillo VA, Stewart RJ, McIntosh JR. Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature. 1995;373:161–164. doi: 10.1038/373161a0. [DOI] [PubMed] [Google Scholar]
- 20.Peskin CS, Oster GF. Force production by depolymerizing microtubules: Load-velocity curves and run-pause statistics. Biophys J. 1995;69:2268–2276. doi: 10.1016/S0006-3495(95)80097-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tao YC, Peskin CS. Simulating the role of microtubules in depolymerization-driven transport: A Monte Carlo approach. Biophys J. 1998;75:1529–1540. doi: 10.1016/S0006-3495(98)74072-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Efremov A, Grishchuk EL, McIntosh JR, Ataullakhanov FI. In search of an optimal ring to couple microtubule depolymerization to processive chromosome motions. Proc Natl Acad Sci USA. 2007;104:19017–19022. doi: 10.1073/pnas.0709524104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shang C, et al. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol Biol Cell. 2003;14:3342–3355. doi: 10.1091/mbc.E02-11-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearson CG, et al. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr Biol. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 25.Joglekar AP, et al. Molecular architecture of kinetochore-microtubule attachment sites is conserved between point and regional centromeres. J Cell Biol. 2008 doi: 10.1083/jcb.200803027. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brouhard GJ, et al. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.