Abstract
MicroRNAs (miRNAs) are an abundant class of small noncoding RNAs that function as negative gene regulators. miRNA deregulation is involved in the initiation and progression of human cancer; however, the underlying mechanism and its contributions to genome-wide transcriptional changes in cancer are still largely unknown. We studied miRNA deregulation in human epithelial ovarian cancer by integrative genomic approach, including miRNA microarray (n = 106), array-based comparative genomic hybridization (n = 109), cDNA microarray (n = 76), and tissue array (n = 504). miRNA expression is markedly down-regulated in malignant transformation and tumor progression. Genomic copy number loss and epigenetic silencing, respectively, may account for the down-regulation of ≈15% and at least ≈36% of miRNAs in advanced ovarian tumors and miRNA down-regulation contributes to a genome-wide transcriptional deregulation. Last, eight miRNAs located in the chromosome 14 miRNA cluster (Dlk1-Gtl2 domain) were identified as potential tumor suppressor genes. Therefore, our results suggest that miRNAs may offer new biomarkers and therapeutic targets in epithelial ovarian cancer.
Keywords: Dlk1-Gtl2 domain, noncoding RNA
Cancer is a disease involving multistep changes in the genome. Studies on cancer genome have so far focused mainly on protein-coding genes, whereas little is presently known on alterations of functional noncoding sequences in cancer (1, 2). MicroRNAs (miRNAs) are endogenous noncoding small RNAs, which negatively regulate gene expression (3–6). In human cancer, miRNAs might function as either oncogenes (7–11) or tumor suppressor genes (12–15). Increasing evidence shows that expression of miRNAs is deregulated in human cancer (1, 2). High-throughput miRNA quantification technologies have provided powerful tools to study global miRNA profiles. It has become progressively obvious that, although the number of miRNAs (≈600) is much smaller than that of the protein-coding genes (≈22,000), miRNA expression signatures reflect more accurately the developmental lineage or tissue origin of human cancers (16). Large-scale studies in human cancer further demonstrated that miRNA expression signatures are associated with specific tumor subtypes and clinical outcomes (16–22). More than half of the miRNAs have been aligned to genomic fragile sites or regions associated with cancers (23), and our group and others have provided evidence that miRNA genes are involved by copying abnormalities in cancer (7, 12, 24). In addition, recent studies suggest that epigenetic alterations might play a critical role in regulating miRNA expression in human cancers (25). Finally, several key proteins in the miRNA biogenesis pathway may be dysfunctional (26) or deregulated in cancer (27–30), which may enhance tumorigenesis (31). Therefore, DNA copy number abnormalities, epigenetic alterations, and/or defects in the miRNA biogenetic machinery might each contribute to miRNA deregulation in human cancer.
Epithelial ovarian cancer (EOC), the most common ovarian malignancy, continues to be the leading cause of death among gynecological malignancies (24). Here, we used integrative genomic approaches to perform a comprehensive analysis of miRNome alterations associated with malignant transformation of the ovarian surface epithelium and/or ovarian tumor stage progression. Our findings indicate that numerous miRNAs are down-regulated in EOC and that this down-regulation can be attributed to genomic copy number loss or more often to epigenetic silencing. Several important miRNA alterations with putative oncogenic or tumor suppressor function were found, including a miRNA cluster in Dlk1-Gtl2 domain that may represent an important therapeutic target in cancer.
Results
microRNA Expression Profiles Classify Malignant from Nonmalignant Ovarian Surface Epithelium.
Most EOC are believed to originate from the ovarian surface epithelium (OSE). Because the OSE represents only a minimal part of the whole normal ovary, whole ovary may not be a suitable normal tissue control for EOC (32). To investigate miRNA alterations associated with OSE malignant transformation, we compared mature miRNA expression profiles in 18 EOC cell lines and four immortalized primary cultured human ovarian surface epithelium (IOSE), using TaqMan miRNA assay. Among 173 miRNAs examined, 160 miRNAs (92.5%) were detected in either IOSEs or EOC cell lines [supporting information (SI) Fig. S1A]. Expression of select miRNAs (mir-15a, mir-30d, mir-182, mir-386, and let-7i) was further confirmed by Northern blot. Unsupervised hierarchical clustering or 3D scaling analysis clearly segregated the two groups of cells (Fig. S1 A and B). There were 35 miRNAs expressed differentially between the EOC and IOSE (P < 0.05). Only four (11.4%, 4/35) were up-regulated, whereas most (88.6%, 31/35) were down-regulated in EOC compared with IOSE lines (Fig. S1C and Dataset S1), including the tumor suppressor miRNAs let-7d (13, 14, 21) and mir-127 (25). Thus, miRNAs are deregulated in EOC and can distinguish malignant from nonmalignant ovarian epithelium. Importantly, most miRNA alterations associated with ovarian epithelial transformation are consistent with down-regulation.
miRNA Down-Regulation in Late-Stage and High-Grade Ovarian Cancer.
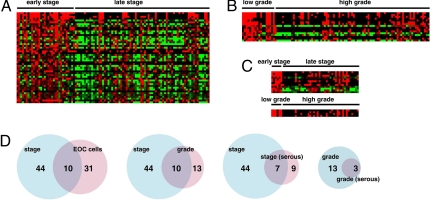
Next, we sought to identify miRNA alterations associated with ovarian cancer progression in vivo. We analyzed 106 primary human ovarian cancer specimens of various stages or grades, using miRNA microarrays. miRNA expression profiles of early-stage (I, n = 25; II, n = 8) and late-stage (III, n = 62; IV, n = 11) EOC were compared by significance analysis of microarrays. Expression of forty-four miRNAs were significantly different between early- and late-stage EOC. Interestingly, all miRNA alterations consisted in down-regulation in late-stage tumors. These alterations included three known tumor suppressors, miRNAs, mir-15a (12), mir-34a, and mir-34b (15, 33–37) (Fig. 1A and Dataset S2). Ten miRNAs were found to be commonly down-regulated in both late-stage (relative to early tumors) and in EOC cells (relative to IOSE cells) (Fig. 1D and Dataset S1 and Dataset S2). In addition, we analyzed miRNA expression differences between the low-grade (0, n = 4; 1, n = 18) and high-grade (2, n = 17; 3, n = 68) EOC. Thirteen miRNAs exhibited significant difference and all were down-regulated in high-grade compared with the low-grade EOC (Fig. 1B and Dataset S3). Ten of 13 miRNAs were commonly down-regulated with stage or grade advancement (Fig. 1D). These results were further validated by stem-loop real-time RT-PCR. From 21 randomly selected miRNAs among the 44 that were down-regulated in late stage, 17 were confirmed to be significantly down-regulated (P < 0.05, early stage n = 30; late stage n = 66) (Fig. S2). Four additional miRNAs were down-regulated in late stage but did not reach statistical significance (Fig. S2). Because the prevalence of histotypes is different among EOC stages (nonserous histotypes prevail among early-stage tumors, whereas serous histotype prevails among late-stage tumors), we tested whether similar miRNA expression differences were detectable in a within-histotype analysis. We analyzed separately serous EOC samples, the most common EOC histotype. Again, we found that miRNA alterations consisted of down-regulation in late-stage or high-grade tumors (Fig. 1C and Dataset S4 and Dataset S5). Taking the primary tumor microarray data and cell line TaqMan data together, we conclude that, during ovarian cancer tumorigenesis and progression, numerous miRNAs are down-regulated by as-yet-unknown mechanisms.
Fig. 1.
Numerous miRNAs are down-regulated in late-stage or high-grade ovarian cancer. (A) Heat map showing the 44 miRNAs significantly down-regulated in late-stage relative to early-stage EOC. (B) Heat map showing the 13 miRNAs significantly down-regulated in high-grade relative to low-grade EOC. (C) Heat map showing the miRNAs significantly down-regulated in late-stage or high-grade serous EOC. (D) Venn diagrams of down-regulated miRNAs in different analyses.
Drosha and Dicer Are Not Deregulated in Ovarian Cancer.
The RNases Drosha and Dicer serve as key regulatory proteins in miRNA biogenesis pathway and their alterations may contribute to widespread miRNA deregulation in cancer (20, 31). Thus, we examined the mRNA expression level of Drosha and Dicer in the same set of EOC samples used for miRNA microarray. There was no significant expression difference in Drosha or Dicer expression between early and late stage EOC (Fig. S3 A and B). Similar levels of Drosha or Dicer were found also in EOC cell lines and IOSE (Fig. S3 C and D). Finally, similar levels of Drosha or Dicer were found in different stage EOC tumors in two independent public cDNA microarrays (Oncomine; Fig. S3 E and F). Next, we analyzed the expression levels of Drosha and Dicer proteins in tissue arrays containing 504 EOC specimens. Immunostaining intensity of these proteins was scored as 1–4, and expression differences between early and late stages were examined by χ2 test. Consistent with the mRNA expression levels, we did not find any significant difference in the expression of Drosha or Dicer between early- and late-stage tumors (Fig. S3 G and H). This result agrees with a recent report showing no substantial down-regulation of the miRNA processing machinery in human tumors (16). To further assess the impact of Drosha or Dicer expression on EOC biology, we performed survival analysis using this tissue array. There was no correlation between the expression levels of either protein and the patient survival (Fig. S4). These results indicate that the observed down-regulation of numerous miRNAs in advanced EOC cannot be attributed to alterations in the key RNases of the miRNA biogenesis pathway.
DNA Copy Number Loss Contributes to the Down-Regulation of miRNAs.
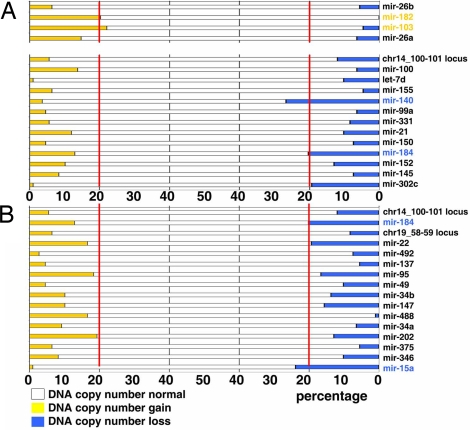
miRNAs are frequently located in cancer-associated regions of the human genome (23), and we have reported that genomic loci containing miRNA genes are frequently altered in human cancers (24). Thus, we examined the chromosomal distribution of miRNAs that were found down-regulated in advanced EOC. Twenty-five of the miRNAs that were down-regulated in late-stage EOC aggregate in <1-Mb clusters in three chromosomes (Fig. S5). These clustered miRNA loci were considered chromosome regions of interest in further aCGH analysis. We first analyzed miRNAs that are deregulated in EOC cell lines. We excluded mir-222, mir-224, and mir-424, which are located in chromosome X (not included in our aCGH platform), and mir-124, which exhibits multiple copies in different genomic loci. The remaining 31 miRNA are located in 17 euchromosomic loci. Two of four (50.0%) loci containing miRNA up-regulated in EOC lines relative to IOSE (mir-182 and mir-103) exhibited amplification (Fig. 2A). Two of 13 (15.3%) loci containing miRNA down-regulated in EOC lines relative to IOSE (mir-140 and mir-184) exhibited deletion (Fig. 2A). Next, we analyzed miRNAs down-regulated in late-stage EOC, again excluding miRNAs located in chromosome X and miRNAs with multiple copies in different genomic loci. This analysis comprised 30 miRNAs located in 16 euchromosomic loci. Two of 16 loci (12.5%, mir-15a and mir-184) were significantly deleted. None of these loci exhibited amplification in late-stage EOC (Fig. 2B).
Fig. 2.
DNA copy number deletions contribute to down-regulation of miRNAs. (A) DNA copy number status of 17 genomic loci containing miRNAs differentially expressed between IOSE cells and EOC cell lines. (Upper) Four up-regulated miRNAs. (Lower) Thirteen down-regulated miRNAs. Red lines indicate the designated cut-off for significant alterations (20%). (B) DNA copy number status of 16 genomic loci containing miRNAs significantly down-regulated in late-stage EOC. Red lines indicate the designated cut-off for significant alterations (20%).
To further confirm that DNA copy number alterations correlate with concordant miRNA deregulation in EOC, we analyzed two representative miRNAs with opposite alterations. We found that locus chr7_129-130, containing mir-182, was amplified in 28.9% of EOC (Fig. S6A). In both primary tumors and cell lines, DNA copy number amplification correlated with miRNA expression (Fig. S6 B and C). Importantly, forced expression of mir-182 in EOC cell line significantly promoted tumor growth in vivo, confirming the role of mir-182 as a putative oncogene (unpublished data). We also analyzed mir-15a, a known tumor suppressor gene (12, 38, 39). mir-15a was deleted in 23.9% of EOC (Fig. S6D). A positive correlation between the deletion of locus chr13_49-50 and reduced expression of mir-15a was found in both primary tumors and cell lines (Fig. S6 E–G). Thus, DNA copy number alteration is one important mechanism of miRNA deregulation in EOC.
Epigenetic Alterations Silence miRNA Expression.
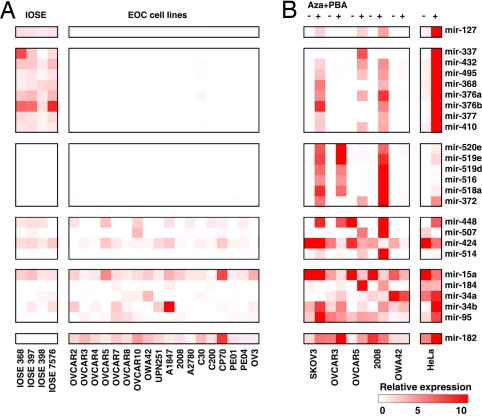
Epigenetic mechanisms play an important role in chromatin remodeling and the regulation of protein-coding genes and miRNA in human cancer (25). Importantly, three genomic loci at chromosomes 14, 19, and X harbor 25 miRNAs down-regulated in EOC (Fig. S5), and these loci may be regulated through imprinting or epigenetic mechanisms (40). Therefore, we analyzed IOSE and EOC cell lines by real-time RT-PCR for expression of 18 from these 25 clustered miRNAs and 5 miRNAs not located in these clusters. mir-127 was chosen as experimental control, because it is epigenetically regulated in human cancer (25). All eight miRNAs located in chromosome 14 cluster (mir-337, mir-432, mir-495, mir-368, mir-376a, mir-376b, mir-377, and mir-419) were expressed in all four IOSE, but in no EOC cell line. None of the six miRNAs located in chromosome 19 cluster (mir-520e, mir-519e, mir-519d, mir-516, mir-518a, and mir-372) were detected in IOSE or EOC cell lines. Finally, three of four miRNAs located in chromosome X cluster (mir-448, mir-507, and mir-424) were expressed in both IOSE and EOC cell lines (Fig. 3A).
Fig. 3.
Epigenetic alterations silence miRNA expression in ovarian cancer. (A) Heat map depicts expression of 24 miRNAs in IOSE cells and EOC cell lines analyzed by real-time RT-PCR. (B) Expression of the same 24 miRNAs in five EOC cell lines and HeLa cells after treatment with demethylating agent 5-aza-2′-deoxycytidine (5-Aza-CdR) and the histone deacetylase inhibitor 4-phenylbutyric acid (PBA) for 6 days.
To investigate whether epigenetic mechanisms are responsible for miRNA down-regulation in EOC, five EOC cell lines were treated with the DNA demethylating agent 5-aza-2′-deoxycytidine (5-Aza-CdR) and the histone deacetylase (HDAC) inhibitor 4-phenylbutyric acid (PBA) (25). Seven of eight miRNAs at the chromosome 14 cluster, six of six miRNAs at the chromosome 19 cluster, two of four miRNAs at the chromosome X cluster, and one of five miRNAs located at other chromosomes were up-regulated by this treatment in at least two cell lines (Fig. 3B). Interestingly, treatment restored the expression of miR-34b, a tumor suppressor miRNA regulated by p53, in all six cell lines. This result was also confirmed by Northern blot (Fig. S7 A and B). Taken together, expression of at least 16 of 44 (36.4%) miRNAs that are down-regulated in late-stage EOC was restored by DNA demethylation or histone deacetylase inhibiting drugs. These in vitro results suggest that epigenetic silencing is an important mechanism contributing to the widespread miRNA down-regulation in late-stage EOC.
MicroRNA Deregulation Affects mRNA Transcripts.
miRNAs can down-regulate mRNA expression by controlling mRNA stability or cleaving target mRNAs. We then asked whether the marked down-regulation of miRNAs observed between early- and late-stage EOC has implications on the transcriptome, i.e., whether it is associated with up-regulation of specific mRNAs. To address this question, we used Affymetrix cDNA microarrays to analyze the genome-wide transcriptional changes between early- and late-stage EOC in samples previously analyzed by miRNA microarray. In total, 76 EOC specimens (8 early-stage and 68 late-stage) were analyzed, and 2,279 transcripts were identified as significantly different between early- and late-stage EOC (P < 0.05). Of these, 1,266 were up-regulated in late-stage EOC. Thus, we collected 948 genes representing the 1,266 up-regulated Affymetrix transcripts and 15,212 annotated genes whose expression was unchanged between early- and late-EOC. Following the method described by Krutzfeld et al. (41), we calculated the number of nonoverlapping occurrences of each possible hexamer in every 3′ UTR sequence of those genes, divided it by the length of the UTR, and applied one-tailed Wilcoxon rank sum to each normalized hexamer count distribution in up-regulated versus unchanged UTRs (Fig. 4 and Fig. S8). Several hexamers were significantly overrepresented in the up-regulated genes (P < 0.001) and in parallel corresponded to the first seed positions (miRNA nucleotides 1–6, 2–7, and 3–8) in miRNAs that were down-regulated in the late-stage EOC. Twelve miRNAs had seed region complementary to the top scoring hexamers (Fig. 4 and Fig. S8B) and were considered significant candidates possibly contributing to the corresponding mRNA up-regulation in late-stage EOC. We also observed an enrichment in up-regulated genes for having at least two hexamers corresponding to the microRNA seed positions (Fig. S8C). Of note, 5 of the top 12 miRNAs identified were located in the chromosome 14 cluster (Dlk1-Gtl2 domain) (42) (Fig. 4, Fig. S5B, and Fig. S8B). This finding indicates that miRNA alterations are not simply “bystander events” during EOC advancement but that they may serve as tumor suppressor genes whose loss mediates oncogenesis.
Fig. 4.
miRNA deregulation affects mRNA transcripts. Histograms of negative natural logarithms of 4,096 P values derived from one-tailed Wilcoxon rank sum test applied to the distributions of each hexamer occurrence in the 3′ UTRs of all up-regulated versus unchanged mRNA transcripts.
miRNA Cluster at Dlk1-Gtl2 Domain Is Commonly Altered in Epithelial Cancers.
Because mir-377, mir-368, and mir-495, three of the eight miRNAs located in the Dlk1-Gtl2 domain of chromosome 14, are also silenced by epigenetic mechanisms in bladder cancer (25), we asked whether down-regulation of miRNAs in this cluster is a common molecular event in human epithelial malignancy. Four miRNAs in this cluster were randomly selected for the analysis in 23 benign breast lesions, 73 invasive ductal carcinomas (IDC), and paired colon cancer and normal colon tissue specimens from 18 patients. All miRNAs were down-regulated in tumors, with all but one (mir-376a) in breast cancer and one (mir-368) in colon cancer reaching statistical significance (Fig. S9). Thus, down-regulation of chromosome 14 miRNA cluster is an event common to many human epithelial tumors. The above results collectively suggest that those eight miRNAs located in chromosome 14 cluster (Dlk1-Gtl2 domain) are commonly regulated and may play a role as tumor suppressor genes in solid tumors, because (i) they are down-regulated in advanced relative to early-stage EOC and in invasive breast and colon cancer, (ii) they are silenced by epigenetic alterations in EOC and bladder cancer (25), and (iii) their down-regulation is associated with up-regulation of numerous potential mRNA targets in late-stage EOC. Importantly, this chromosome region has been identified as a cancer susceptibility locus in mouse (43). Therefore, we further analyzed the putative mRNA targets of these eight miRNAs. mRNA targets identified with the above method for each of these miRNAs are listed in Dataset S6. Gene ontology (GO) analysis (18) revealed that these miRNAs are highly associated with two important biological processes implicated in human carcinogenesis, namely cell cycle regulation and immune response (Dataset S7).
Down-Regulation of miRNA Cluster at Dlk1-Gtl2 Domain Is Associated with Poor Survival.
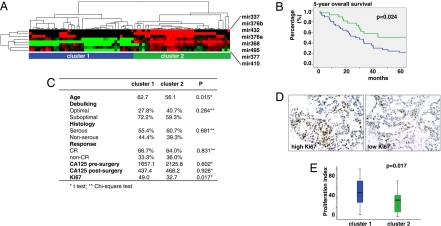
We asked whether deregulation of the miRNAs at the Dlk1-Gtl2 domain affects tumor behavior at an advanced stage. Nonsupervised clustering of eight Dlk1-Gtl2 domain miRNA expression signature classified 73 late-stage EOC in two distinct clusters, cluster 1, with global low expression of these miRNAs (n = 38), and cluster 2, with relatively global high expression (n = 35) (Fig. 5A). Sixty-four tumors with histological sections and clinical follow-up were available for analysis. Patients in cluster 1 were at significantly greater risk with shorter 5-year survival than those in cluster 2 (P = 0.024, log-rank test, Fig. 5B). Because miRNAs located at Dlk1-Gtl2 domain appear to regulate mRNAs involved in the control of cell cycle, we examined the tumor proliferation index (percentage of Ki67-positive cells in tumor islets). Tumors in cluster 1 exhibited significantly higher proliferation index (49.0) than cluster 2 (32.7, P = 0.017, Fig. 5 C–E). Thus, down-regulation of miRNAs located at Dlk1-Gtl2 domain is associated with higher tumor proliferation and shorter patient survival.
Fig. 5.
Down-regulation of miRNA cluster at the Dlk1-Gtl2 domain is associated with poor survival. (A) Nonsupervised clustering of the eight Dlk1-Gtl2 domain miRNA expression signatures classifies 73 late-stage EOC in two distinct clusters (cluster 1, n = 38; cluster 2, n = 35). (B) Five-year survival of patients with advanced stage EOC whose tumors belong to cluster 1 (blue) or cluster 2 (green). (C) Summary of clinicopathologic characteristics of patients in the two clusters. (D) Examples of high and low proliferation index based on Ki67 immunohistochemistry staining in late-stage EOC. (E) Summary of proliferation index in tumors from the two clusters.
Discussion
According to recent high-throughput studies, global expression of miRNAs is seemingly deregulated in most cancer types (1, 16–20). Interestingly, miRNA expression may be widely down-regulated in human tumors relative to normal tissues, as revealed by bead-based flow cytometry (16) or miRNA microarray (44). However, other microarray studies reported a tumor-specific mixed pattern of down-regulation and up-regulation of select miRNA genes (17–19). The choice of control samples may therefore be critical in the interpretation of those results. For example, normal ovaries are mainly comprised of stroma and the surface epithelium represents only a minimal part; thus, the whole ovary may not serve as an optimal control for EOC studies. We accordingly compared EOC cell lines with ovarian surface epithelial cells to investigate the miRNA alterations associated with OSE malignant transformation. Our findings indicate that the miRNome is largely down-regulated in association with OSE malignant transformation and EOC progression, which is in agreement with recent reports (45, 46). This result further suggests that certain miRNAs may function as tumor suppressor genes and justifies why abrogation of miRNAs at large may in fact be a hallmark of human cancers (16).
DNA copy number abnormalities (23, 24), epigenetic alterations (25), mutations (17), transcriptional deregulation (15), and defective miRNA biogenesis pathway (31) may contribute to the miRNA deregulation in human cancer. In EOC, we showed that: (i) Drosha and Dicer, key proteins in miRNA biogenesis pathway, are not altered and, thus, are unlikely to contribute to the miRNA deregulation between early- and late-stage EOC; (ii) deletions occur in up to 15% of genomic loci harboring miRNAs that are down-regulated, suggesting that genomic loss contributes to miRNA down-regulation in EOC; and (iii) at least one-third of down-regulated miRNAs may be silenced by epigenetic alterations. For example, miR-15a was down-regulated and its DNA copy number was deleted in EOC. miR-34b, a tumor suppressor miRNA regulated by p53 (15, 33–37), was also down-regulated in EOC but because of epigenetic silencing. Complementary mechanisms, including transcriptional regulation, may cooperate in miRNA deregulation. For example, mir-34b is regulated by p53, and p53 loss could cooperate with epigenetic mechanisms to down-regulate mir-34b. However, different mechanisms may regulate miRNA in opposite directions. For example, genomic gain may increase mir-182 expression in EOC, but mir-182 is silenced epigenetically in some EOC cell lines (Fig. 5B) and bladder cancer (25). Thus, final expression will depend on the net effect of these mechanisms. In addition, this study carried out the very first analysis to link miRNA microarray to cDNA microarray data to provide, through seed sequences, molecular evidence that miRNA down-regulation can indeed result in specific genome-wide transcriptional up-regulation.
Finally, a putative tumor suppressor miRNA cluster located at chromosome 14 (Dlk1-Gtl2 domain) was identified. This chromosome region is a cancer susceptibility locus in the mouse (43) and AAV integration in this location induced hepatocellular carcinoma (47). Eight miRNAs located in this cluster are of particular interest, because they were suppressed in EOC cell lines in advanced EOC and invasive breast and colon cancer, they were silenced by epigenetic mechanisms, and they were predicted to up-regulate a large number of mRNA transcripts in late stage EOC that, by GO analysis, are implicated in cancer. Some of these miRNAs are also down-regulated in bladder cancer (25). miRNAs in this chromosome region are only expressed from a maternally inherited chromosome and their imprinted expression is regulated by an intergenic germ line-derived differentially methylated region located ≈200 kb upstream of the miRNA cluster (42). Importantly, tumors with lower expression of these eight miRNAs were associated with higher proliferation index and significantly shorter survival. At this point, the function of this miRNA cluster is largely unknown, but it may play a critical role in embryonic development (42, 48, 49). Our data suggest that miRNAs in this cluster function as tumor suppressor genes. Further work is required to understand their function and potential for cancer therapy.
Materials and Methods
Patients and Specimens.
The ovarian cancer (miRNA microarray, n = 106; aCGH, n = 109; Affymetrix cDNA microarray, n = 76; tissue array, n = 504; and qPCR validation, n = 96) and breast cancer (n = 96) specimens were collected at the University of Pennsylvania; the University of Turin, and the University of Helsinki. Detailed information is provided in SI Methods. The colon cancer and paired normal control specimens (n = 18 pairs) were provided by the cooperative human tissue network (Cooperative Human Tissue Network) and the Department of Surgical Oncology, Okayama University.
TaqMan miRNA Assay.
Expression of mature miRNAs was analyzed by TaqMan miRNA Assay (Applied Biosystems). Detailed information is provided in SI Methods.
miRNA Microarray.
miRNA microarray was performed on the microarray chip (OSU_CCC version 3.0; OSU). Detailed information is provided in SI Methods.
cDNA Microarray.
cDNA microarray was performed on the human U133 + 2.0 GeneChip (Affymetrix). Detailed information is provided in SI Methods.
Array-Based Comparative Genomic Hybridization (aCGH).
BAC clones included in the 1-Mb array platform were described in ref. 24. Detailed information is provided in SI Methods.
Tissue Microarray.
The tissue microarray was constructed as described. A total of 504 specimens were printed in the array slides. Detailed information is provided in SI Methods.
5-Aza-CdR and PBA Treatment.
Treatment was preformed as descried by Saito (25). Briefly, cells were seeded at 5 × 105 cells per T75 flask 24 h before treatment with 5-Aza-CdR (3 μM; Sigma–Aldrich) and/or PBA (3 mM; Sigma–Aldrich). 5-Aza-CdR was removed after 24 h, whereas PBA was continuously administered by replacing the medium containing PBA every 24 h for 6 days.
Bioinformatic Analysis.
Detailed information for miRNA target prediction and gene ontology analysis is provided in SI Methods.
Statistics.
Statistical analysis was performed by using the SPSS statistics software package (SPSS). All results were expressed as mean ± SD, and P < 0.05 was used for significance. Kaplan–Meier curves were used to estimate 5-year rates and were compared with the use of log-rank statistics.
Supplementary Material
Acknowledgments.
We thank Vassilis Altmazoglou and Trias Thiraiou for the mapping of the Affymetric Gene ID to the Ensembl Gene ID and the initial calculation on the hexamer distribution. We thank the following investigators who developed public microarray analysis software: TM4 developed was by Dr. John Quackenbush (Harvard School of Public Health, Boston); BRB ArrayTools were developed by Drs. Richard Simon and Amy Peng Lam (National Cancer Insitute, Rockville, MD); GenePattern was developed by Dr. Jill P. Mesirov (Broad Institute, Cambridge, MA); and Cluster software was developed by Drs. Michael Eisen (Lawrence Berkeley National Laboratory, Berkeley, CA) and David Botstein (Princeton University, Princeton). We thank Dr. Nelly Auersperg (University of British Columbia, Vancouver, BC, Canada) for IOSE and access to the Canadian Ovarian Tissue Bank. This work was supported by a grants from the Ovarian Cancer Research Fund (G.C. and L.Z.), National Cancer Institute ovarian Specialized Programs of Research Excellence Grant P01-CA83638 Career Development Award (to L.Z.), American Cancer Society Grant IRG-78-002-30 (to L.Z.), the Mary Kay Ash Charitable Foundation (L.Z.), National Science Foundation Grant DBI-0238295 (to A.G.H.), and National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases Grant R01-DK056645 (to A.K.R. and C.N.J.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801615105/DCSupplemental.
References
- 1.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 2.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 5.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 6.Zamore PD, Haley B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 7.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Donnell KA, et al. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 9.Costinean S, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voorhoeve PM, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 11.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 12.Calin GA, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 17.Calin GA, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 18.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 19.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cummins JM, et al. The colorectal microRNAome. Proc Natl Acad Sci USA. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shell S, et al. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA. 2007;104:11400–11405. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu L, et al. Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Cancer Res. 2007;67:10117–10122. doi: 10.1158/0008-5472.CAN-07-2544. [DOI] [PubMed] [Google Scholar]
- 23.Calin GA, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saito Y, et al. Specific activation of microRNA-127 with down-regulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 26.Thomson JM, et al. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muralidhar B, et al. Global microRNA profiles in cervical squamous cell carcinoma depend on Drosha expression levels. J Pathol. 2007;212:368–377. doi: 10.1002/path.2179. [DOI] [PubMed] [Google Scholar]
- 28.Chiosea S, et al. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345–2350. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- 29.Chiosea S, et al. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–1820. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karube Y, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar MS, et al. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 32.Zorn KK, et al. Choice of normal ovarian control influences determination of differentially expressed genes in ovarian cancer expression profiling studies. Clin Cancer Res. 2003;9:4811–4818. [PubMed] [Google Scholar]
- 33.Bommer GT, et al. p53-Mediated Activation of miRNA34 Candidate Tumor-Suppressor Genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 34.Chang TC, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raver-Shapira N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 36.Tarasov V, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 37.Corney DC, et al. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 38.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raveche ES, et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice, Blood. 2007;109:5079–5086. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Royo H, Bortolin ML, Seitz H, Cavaille J. Small non-coding RNAs and genomic imprinting. Cytogenet Genome Res. 2006;113:99–108. doi: 10.1159/000090820. [DOI] [PubMed] [Google Scholar]
- 41.Krutzfeldt J, et al. Silencing of microRNAs in vivo with “antagomirs”. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 42.Seitz H, et al. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 2004;14:1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sevignani C, et al. MicroRNA genes are frequently located near mouse cancer susceptibility loci. Proc Natl Acad Sci USA. 2007;104:8017–8022. doi: 10.1073/pnas.0702177104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Visone R, et al. Specific microRNAs are down-regulated in human thyroid anaplastic carcinomas. Oncogene. 2007;26:7590–7595. doi: 10.1038/sj.onc.1210564. [DOI] [PubMed] [Google Scholar]
- 45.Iorio MV, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 46.Yang H, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 47.Donsante A, et al. AAV vector integration sites in mouse hepatocellular carcinoma. Science. 2007;317:477. doi: 10.1126/science.1142658. [DOI] [PubMed] [Google Scholar]
- 48.Lin SP, et al. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet. 2003;35:97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- 49.Lin SP, et al. Differential regulation of imprinting in the murine embryo and placenta by the Dlk1-Dio3 imprinting control region. Development (Cambridge, UK) 2007;134:417–426. doi: 10.1242/dev.02726. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.