In December 2003, Giot et al. (1) published a systematic investigation of the protein interaction network—the interactome—of Drosophila melanogaster. Giot et al. produced a draft map of 7,048 proteins and 20,405 interactions, which they then refined to “a higher confidence map of 4,679 proteins and 4,780 interactions.” The magnitude of the undertaking led to the study being lauded as the “dawn of systems biology” in a number of commentaries and news releases. Giot et al.'s study was preceded and followed by a number of investigations of the interactomes of other species, ranging from bacteria to humans (2–5). However, none of these studies was able to provide an estimate of the actual size of the interactome being sampled. In a systematic statistical study published in this issue of PNAS, Stumpf et al. (6) provide convincing estimates of the interactome size of four organisms, including humans.
Stumpf et al. (6) estimate that the human interactome comprises ≈25,000 proteins and on the order of 650,000 interactions. These numbers provide a sobering view of where we stand in our cataloging of the human interactome. At present, we have identified <0.3% of all estimated interactions among human proteins. We are indeed at the dawn of systems biology.
The sparse sampling of the human interactome should make researchers distrustful of the numerous studies reporting global analysis of human protein interaction networks. As Stumpf et al. (6) stress, the actual size of the interactome may be one of the only global characteristics that can be estimated in an unbiased manner from small, biased samples. This is particularly true of the human interactome: Although the library of probes in most studies is likely to be unbiased, the set of targets is likely selected on the basis of expectations of importance for development, regulation, or disease. The consequences of this sampling scheme are clearly visible in the multistar structure of protein interactions networks, as demonstrated by Guimerà et al. (7), and they should make one suspicious of broad claims.
Stumpf et al.'s (6) analysis also reveals that the human interactome is nearly 10 times larger than that of D. melanogaster and 3 times larger than that of Caenorhabditis elegans. As the authors state, “interactome sizes are consistent with biological intuition about complexity of eukaryotic organisms” (6). Although this is surely reassuring to those needing supporting evidence for the greater complexity of Homo sapiens, it may be placing emphasis on the wrong concern. It takes no more than common sense to realize that humans are more complex organisms than fruit flies or yeasts. The fact that a coarse measure of complexity, such as gross number of base pairs/genes/proteins, does not capture the clear qualitative difference in complexity between humans and those organisms merely reveals that there are still a large number of open questions about how biological complexity emerged and how it is implemented. Indeed, the big, fundamental question driving systems biology must be thus: Which molecular components and organizational motifs among those components enable the emergence of different levels of biological complexity?
The set of targets is selected on the basis of expectations.
To answer the question above, we will need to address another question of equal importance: How do we make sense of the “seas” of biological data we are gathering by high-throughput methods? (8). The complexity of the data we are now able to gather makes it not at all surprising that our understanding of biomedical systems has fallen behind our ability to gather new data. Our brains likely evolved the capacity to process, in a meaningful manner, only a handful of components, not the tens of thousands we find in biological systems. However, it is now clear that reductionist approaches alone will not enable us to solve many of today's most important biomedical questions. Understanding the folding of a single protein is not going to bring deep insights into the origins or progression of cancer, just as unveiling the working of a single neuron cannot provide an understanding of consciousness.
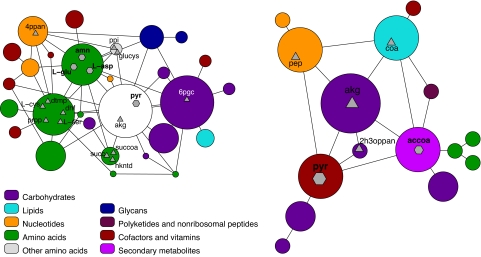
A saving grace may be the fact that biological complexity has a hierarchical organization: organism → organ → tissue → cell → pathway → motif → molecule. This hierarchical structure is analogous to the structure of geopolitical entities: continents → countries → states → regions → counties → localities → neighborhoods → buildings. Like any organizational scheme, the way geopolitical entities are classified is not always straightforward or free of information loss. However, the classification is extraordinarily powerful in enabling users of the information to easily locate even the components relevant only at the lowest scale. The reason for this ease-of-use is the fact the hierarchical representations are scalable: The representation is able to extract the information that is most relevant at the scale of interest (Fig. 1).
Fig. 1.
Mapping the metabolism of Escherichia coli. (Left) Map of a metabolic network of E. coli, which comprises 507 metabolites and 718 connections (11). The area of the circles is proportional to the number of metabolites in the corresponding module. The hexagons indicate connector hub metabolites, and the triangles indicate satellite connector metabolites. (Right) Map of the module containing pyruvate. The smaller symbols and fonts indicate roles at the second level in the hierarchy. 4ppan, d-4′-phosphopantothenate; amn, ammonia; L-glu, l-glutamate; L-asp, l-aspartate; ppi, diphosphate; glucys, γ-l-glutamyl-l-cysteine; L-cys, l-cysteine; L-ser, l-serine; dtmp, dTMP; dhf, 7,8-dihydrofolate; prpp, 5-phospho-α-d-ribose 1-diphosphate; pyr, pyruvate; akg, 2-oxoglutarate; succ, succinate; succoa, succinyl-CoA; hkntd, 2-hydroxy-6-ketononatrienedioate; 6pgc, 6-phospho-d-gluconate; pep, phosphoenolpyruvate; 2h3oppan, 2-hydroxy-3-oxopropanoate; accoa, acetoacetyl-CoA; coa, CoA. Figure courtesy of R. Guimerà and M. Sales-Pardo (both at Northwestern University).
These facts prompt the need to develop a cartography for complex biological networks (9). Such a cartography would aim to do what geopolitical cartography did for the representation of geopolitical information. The cartographic approach is based on two core assumptions (9, 10). The first assumption is that the nodes in a network can be grouped into modules, thus enabling a simplified description of the network. It is important to note that despite much work on clustering and the widespread use of hierarchical clustering methods, there was, until recently, no procedure that enabled one to simultaneously assess whether a network is organized in a hierarchical fashion and to identify the different levels in the hierarchy in an unsupervised manner. Indeed, many methods, such as hierarchical clustering, yield a hierarchical tree even for networks with no internal structure (11). Work by numerous researchers on the detection of modular structure of complex networks (12), has recently culminated in a new method that is able to determine the hierarchical structure of complex networks of arbitrary type (11).
The second core assumption of the cartographic approach is that one can classify the nodes comprising a network into a small number of system-independent “universal roles.” Guimerà and Amaral (9) proposed a classification scheme that rests on the expectation that the nodes in a network are connected according to the specific purpose they fulfill. Specifically, the role of a node is defined according to (i) how many connections it has and (ii) to what degree the node is a connector of different modules. Guimerà and Amaral (9) defined four main types of roles: hub connectors, which have many connections to both other nodes in their module and nodes in other modules; provincial hubs, which have many connections but only to nodes inside their module; satellite connectors, which have few connections but act as bridges between modules; and peripheral nodes, which have few connections, mostly to nodes inside their module.
To demonstrate the power of this cartographic perspective, Guimerà and Amaral (9) studied the overall organization of the cellular metabolisms of twelve organisms (13, 14, 15). They found that ≈90% of the metabolites in these organisms are classified as peripheral nodes, suggesting a very weak signal-to-noise ratio. The important metabolites are a small fraction of all metabolites, thus limiting information loss when coarse-graining.
The graphical representations of the protein networks in the literature make very clear the problem of information overload we are already experiencing. Stumpf et al. (6) reveal to us, in no uncertain terms, that those images capture no more than a tiny fraction of the system. This should convince all parties involved of the need to develop coarse-grained representations of biological systems. The reward of such an undertaking is clear: With these maps at their fingertips, researchers, physicians, and educators will be able to navigate the seas of biological data to easily locate, and ultimately manipulate, biological systems of interest (16).
Acknowledgments.
I gratefully acknowledge the support of the Keck Foundation and of a National Institutes of Health/National Institute of General Medical Sciences K-25 Award.
Footnotes
The author declares no conflict of interest.
See companion article on page 6959.
References
- 1.Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 2.Uetz P, et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 3.Rain JC, et al. The protein–protein interaction map of Helicobacter pylori. Nature. 2001;409:211–216. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 4.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 5.Rual JF, et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 6.Stumpf PH, et al. Estimating the size of the human interactome. Proc Natl Acad Sci USA. 2008;105:6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guimerà R, Sales-Pardo M, Amaral LAN. Classes of complex networks defined by role-to-role connectivity profiles. Nat Phys. 2007;3:63–69. doi: 10.1038/nphys489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pennisi E. How will big pictures emerge from a sea of biological data? Science. 2005;309:94. doi: 10.1126/science.309.5731.94. [DOI] [PubMed] [Google Scholar]
- 9.Guimerà R, Amaral LAN. Functional cartography of complex metabolic networks. Nature. 2005;433:895–900. doi: 10.1038/nature03288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guimerà R, Amaral LAN. Cartography of complex networks: Modules and universal roles. J Stat Mech Theor Exp. 2005 doi: 10.1088/1742-5468/2005/02/P02001. article no. P02001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sales-Pardo M, Guimerà R, Moreira AA, Amaral LAN. Extracting the hierarchical structure of complex systems. Proc Natl Acad Sci USA. 2007;104:15224–15229. doi: 10.1073/pnas.0703740104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danon L, et al. Comparing community structure identification. J Stat Mech Theor Exp. 2005 article no. P09008. [Google Scholar]
- 13.Lee SY, Papoutsakis ET, editors. Metabolic Engineering. New York: Marcel Dekker; 1999. [Google Scholar]
- 14.Schuster S, Fell DA, Dandekar T. A general definition of metabolic pathways useful for systematic organization and analysis of complex metabolic networks. Nat Biotechnol. 2000;18:326–332. doi: 10.1038/73786. [DOI] [PubMed] [Google Scholar]
- 15.Palsson B. Systems Biology–Properties of Reconstructed Networks. Cambridge, UK: Cambridge Univ Press; 2006. [Google Scholar]
- 16.Apic G, Ignjatovic T, Boyer S, Russell RB. Illuminating drug discovery with biological pathways. FEBS Lett. 2005;579:1872–1877. doi: 10.1016/j.febslet.2005.02.023. [DOI] [PubMed] [Google Scholar]