Abstract
Background
Comparative genome analyses of parasites allow large scale investigation of selective pressures shaping their evolution. An acute limitation to such analysis of Plasmodium falciparum is that there is only very partial low-coverage genome sequence of the most closely related species, the chimpanzee parasite P. reichenowi. However, if orthologous genes have been under similar selective pressures throughout the Plasmodium genus then positive selection on the P. falciparum lineage might be predicted to some extent by analysis of other lineages.
Principal Findings
Here, three independent pairs of closely related species in different sub-generic clades (P. falciparum and P. reichenowi; P. vivax and P. knowlesi; P. yoelii and P. berghei) were compared for a set of 43 candidate ligand genes considered likely to be under positive directional selection and a set of 102 control genes for which there was no selective hypothesis. The ratios of non-synonymous to synonymous substitutions (dN/dS) were significantly elevated in the candidate ligand genes compared to control genes in each of the three clades. However, the rank order correlation of dN/dS ratios for individual candidate genes was very low, less than the correlation for the control genes.
Significance
The inability to predict positive selection on a gene in one lineage by identifying elevated dN/dS ratios in the orthologue within another lineage needs to be noted, as it reflects that adaptive mutations are generally rare events that lead to fixation in individual lineages. Thus it is essential to complete the genome sequences of particular species of phylogenetic importance, such as P. reichenowi.
Introduction
Identifying genes under positive directional selection can help understand how parasites adapt to new survival or reproductive challenges. The dN/dS ratio (non-synonymous substitutions per non-synonymous site divided by synonymous substitutions per synonymous site) is commonly applied to scan for evidence of positive selection in comparative genomic analysis [1], [2]. Analyses of polymorphism among genome sequences of the human malaria parasite P. falciparum [3]–[5], and divergence between P. falciparum and the partially available genome sequence of the chimpanzee parasite P. reichenowi [3] show elevated dN/dS ratios in genes encoding membrane and exported proteins (considered to be under positive selection), as well as genes that are expressed at low abundance or at only one stage of the life cycle (considered to be under relaxed negative selection). However, the incompleteness of the P. reichenowi genome sequence (available sequence reads aligned to only ∼ 42% of the P. falciparum 3D7 genome sequence) means that most loci could not be effectively analysed for inter-specific divergence [3], so most signatures of positive directional selection have not yet been discriminated.
Pairwise analyses with other malaria parasite species may also identify loci under positive selection. However, given the great evolutionary distance between many of the species, such as between P. falciparum and the rodent parasite P. yoelii [6], studies of pairwise dN/dS suffer from too high a sequence divergence, causing synonymous substitutions to be saturated and making estimates of dN/dS rate ratios unreliable. Analyses of closely related species are preferable, and pairwise dN/dS analysis among the genomes of the rodent malaria parasites, P. yoelii, P. berghei and P. chabaudi [7], showed a similar overall trend to the falciparum-reichenowi analysis, with putative membrane proteins displaying higher dN/dS values than other genes. Could the results of that analysis (or analysis of other closely related species pairs such as P. vivax and P. knowlesi) be extrapolated to P. falciparum genes for which P. reichenowi orthologous sequences are not available? This study tests whether signatures from one clade of the Plasmodium genus can be used to predict those in other clades. The distributions of dN/dS values are compared for sets of orthologous loci in three phylogenetically independent species pairs, investigating a set of 43 candidate genes that are considered likely to be under positive selection and a set of 102 control genes for which there is no selective hypothesis.
Results and Discussion
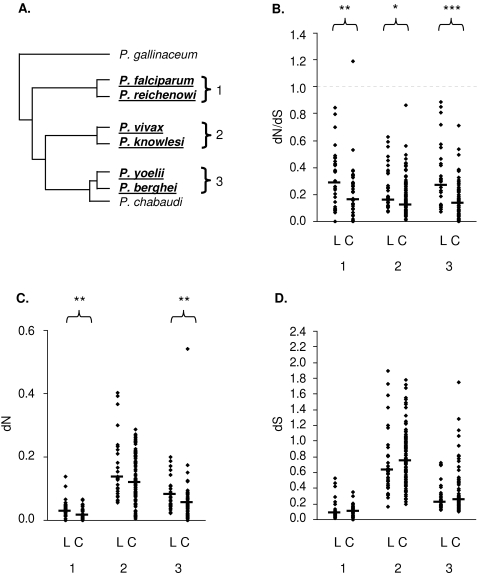
For each of the 43 candidate ligand genes analysed, inter-specific dN/dS ratios are shown for each of the three closely related species pairs, P. falciparum / P. reichenowi, P. vivax / P. knowlesi, and P. yoelii / P. berghei (Table 1, further details in table S1). To test whether this candidate ligand gene dataset is enriched in genes under positive selection, dN/dS values were compared with the control gene dataset (table S2) for each species pair (Fig. 1A) using Wilcoxon's rank sum test. For all three species pairs the median dN/dS ratio was significantly greater in the candidate ligand gene set than in the control set (falciparum-reichenowi, P = 0.0084; vivax-knowlesi, P = 0.0175; yoelii-berghei, P = 0.0003) (Fig. 1B). This was also seen for dN values (Fig. 1C), though not for dS (Fig. 1D), indicating a signature of positive selection on non-synonymous mutations leading to elevated dN/dS values in a proportion of the candidate ligand genes. Relaxed selective constraint could also result in elevated dN/dS, although there is no reason to expect that the candidate ligand genes should be under any less selective constraint than the control set of genes to maintain protein structure and function.
Table 1. A set of 43 candidate ligand gene loci with dN/dS ratios for three phylogenetically independent Plasmodium species pairs (Pf/Pr, Pv/Pk and Py/Pb).
| Pf locus ID | Gene product | Evidence for ligand role | References | Pf/Pr | Pv/Pk | Py/Pb |
| PFB0310c | MSP4 | Da, SEm | [20], [21] | 0.84 | 0.48 | -n |
| PFB0305c | MSP5 | SEm | [21], [22] | 0.80 | 0.59 | -n |
| PF13_0201 | TRAP | B, IIA, Db, AEs, SEs | [23]–[25] | 0.70 | 0.35 | 0.72 |
| MAL13P1.60 | EBA140 | B, IIA, AEm | [26]–[28] | 0.57 | 0.42 | -n |
| PF10_0352 | MSP11 | SEm | [29] | 0.49 | -n | -n |
| PF11_0486 | MAEBL | B, IIA, Db, SEs, AEm | [30]–[32] | 0.48 | 0.37 | 0.30 |
| PFA0125c | EBA181 | B, AEm | [33] | 0.47 | -n | -n |
| PFE0080c | RAP2 | AEm | [34], [35] | 0.44 | -n | -n |
| PF10_0302 | P28 | IIA, Db, SEo | [36], [37] | 0.43 | -n | 0.57 |
| PF13_0248 | P47 | SEg | [38] | 0.42 | 0.12 | 0.55 |
| PFD1150c | RH4 | AEm | [39]–[41] | 0.42 | -n | -n |
| PFD0210c | P36 | Db | [42] | 0.42 | 0.18 | 0.32 |
| PF10_0303 | P25 | IIA, Db, SEo | [36], [37] | 0.38 | 0.45 | 0.81 |
| PF14_0102 | RAP1 | IIA, AEm | [34], [35], [43] | 0.37 | 0.19 | 0.85 |
| PFF0615c | Pf12 | SEm | [44] | 0.37 | 0.14 | 0.12 |
| PFF0995c | MSP10 | Da, AEm | [21], [45] | 0.30 | 0.38 | -p |
| PF11_0344 | AMA1 | B, IIA, Da, AEm/SEm, AEs/SEs | [46]–[48] | 0.30 | 0.14 | 0.30 |
| PFL0800c | celTOS | Db, AEo | [49] | 0.26 | 0.63 | -n |
| PF10_0344 | GLURP | SEm | [50] | 0.25 | -n | -n |
| PFE0395c | Pf38 | SEm | [44] | 0.24 | 0.11 | 0.25 |
| PFI1730w | CLAG9 | Dc, AEm | [51], [52] | 0.24 | -n | -n |
| PFC0640w | CTRP | Db, AEo | [53], [54] | 0.24 | 0.25 | 0.34 |
| PFC0210c | CSP | B, SEs | [55]–[57] | 0.20 | 0.56 | 0.41 |
| PFI1445w | RhopH2 | AEm | [58] | 0.18 | 0.07 | 0.10 |
| PF13_0247 | P48/45 | Dd, SEg | [59] | 0.14 | 0.15 | 0.21 |
| PFI0265c | RhopH3 | Da, AEm | [60], [61] | 0.11 | -n | 0.32 |
| PFL2510w | CHT1 | Db, AEo(secreted) | [62], [63] | 0.10 | 0.14 | 0.13 |
| PFE0075c | RAP3 | AEm | [34] | 0.09 | -n | 0.89 |
| PFL0870w | PTRAMP | AEm | [64] | 0.08 | 0.18 | 0.10 |
| MAL7P1.208 | RAMA | B, Da, AEm | [21], [65] | 0.08 | 0.23 | 0.47 |
| PFB0405w | P230 | B, IIA, SEg | [66], [67] | 0.08 | 0.11 | 0.23 |
| PFC0120w | CLAG3.2 | AEm | [68] | 0.07 | -n | -n |
| PFB0570w | SPATR | B, IIA, SEs | [69] | 0.001 | 0.12 | 0.09 |
| PF08_0003 | TryThrA | IES | [70] | -n | -p | 0.10 |
| PF14_0040 | SOAP | Db, AEo | [71] | -n | 0.56 | -n |
| PF13_0338 | Pf92 | Da, SEm | [21], [44] | -n | 0.27 | -n |
| PF08_0136b | WARP | AEo | [72] | -n | 0.26 | 0.43 |
| PFD0215c | P36p | Db, SEs | [42] | -n | 0.25 | 0.30 |
| PFE0120c | MSP8 | SEm(in P. yoelii) | [73] | -n | 0.18 | 0.12 |
| PFI1145w | PLP3/MAOP | Db, AEo | [74] | -n | 0.17 | 0.13 |
| PFL1385c | MSP9 | B, SEm | [75], [76] | -n | 0.17 | 0.07 |
| PFD0240c | Pf41 | SEm | [44] | -n | 0.16 | 0.30 |
| PFC0420w | CDPK3 | Db, AEo | [77], [78] | -n | 0.08 | 0.18 |
B = binding assay. IIA = invasion inhibition assay. D = gene disruption experiment which either (a) could not produce viable parasites in asexual culture; (b) reduced or abolished the traversal of cell membranes or tissue layers; (c) abolished receptor binding; or (d) reduced fertilization. SE/AE/IES = surface/apical/infected erythrocyte surface expression at (s) sporozoite, (m) merozoite, (g) gametocyte, or (o) ookinete stage. -n unambiguous orthologues could not be identified in one or both species; -p orthology could not be resolved among alternative possible orthologues. For each gene a maximum of three references are given. Twelve other candidate ligand loci could not be analysed due to complex sequence evolution (see Methods).
Figure 1. Genetic divergence among three pairs of Plasmodium species.
A. Schematic representation of the phylogenetic relationship between sequenced Plasmodium genomes. Three pairs of closely related species (falciparum-reichenowi, vivax-knowlesi and yoelii-berghei) used for analysis are labelled clade 1, 2 and 3, respectively. (The phylogenetic position of P. gallinaceum in relation to the other species is not yet confirmed and awaits full genomic analysis, but is either an outgroup as illustrated here [10], [11] or more closely related to the falciparum-reichenowi clade). B. The distribution of dN/dS for candidate ligand genes and control genes (labelled ‘L’ and ‘C’) between species of each clade defined in panel A. Sample sizes were: clade 1, L = 33, C = 37; clade 2, L = 32, C = 92; clade 3, L = 29, C = 70. Asterisks indicate a significant difference between gene datasets by Wilcoxon's rank sum test (*<0.05, **<0.01, ***<0.001). C. The distribution of dN for the same loci. One extreme value (PY05686 vs. PB000528.03.0, dN = 8.06) is not shown. D. The distribution of dS for the same loci. Two extreme values (PY05686 vs. PB000528.03.0, dS = 45.69; PY02848 vs. PB100183.00.0, dS = 108.79) are not shown.
It should be noted that analysis of any single one of these genes in isolation would not lead to a strong conclusion of positive selection, since none showed a dN/dS value >1. Inter-specific dN/dS values for whole genes are hardly ever >1 even when positive selection occurs, due to the effect of negative background selection on many sites within most genes [1], [2], so comparison of relative dN/dS values across sets of genes is a more sensitive way of scanning for evidence of positive selection than searching for individual values above 1 or any other arbitrary cut off.
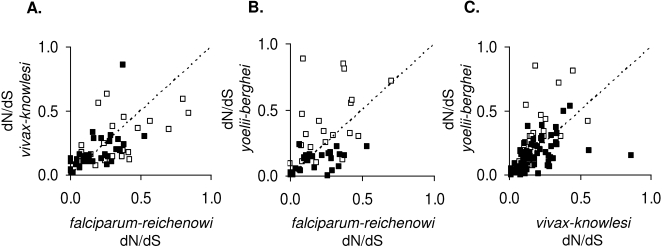
To assess the predictive power of dN/dS across the Plasmodium genus, rank correlations (Spearman's ρ = rSp) were applied to test whether similar relative selective forces operate on orthologous genes in different species. Table 2 shows the correlation of dN/dS, dN and dS indices for all genes among the three different species pairs. Pairwise scatterplots of dN/dS values are shown in Fig. 2. The predictive power is quantified by r2Sp which represents the amount of variability in one axis which can be explained by variability in the other. dN/dS was significantly, though poorly, positively correlated between independent species pairs for both candidate ligand genes and control genes. Correlations were greater for dN than for dS, supporting the idea that selection affects the correlations while synonymous substitutions are mostly stochastic. However, the predictive power of dN/dS for one species pair on another is lower for candidate ligand genes (25 %, 21 % and 31 % for Pf/Pr versus Pv/Pk, Pf/Pr versus Py/Pb, and Pv/Pk versus Py/Pb respectively) than for control genes (55 %, 35 % and 44 % for the respective three comparisons). This indicates that the correlation is not improved by positive selection but is actually made worse. Discrete processes of positive selection will have occurred in different species lineages, against a background of selective constraint that varies among genes in a manner that is apparently more homogeneous between different lineages.
Table 2. Spearman's rank correlation (rSp) of pairwise sequence divergence estimates for orthologous loci among different species pairs.
| Species pairs compared | Gene dataset | N | Index | rSp | r2Sp | |
| falciparum-reichenowi versus vivax-knowlesi | Ligand | 23 | dN/dS | 0.50 | 0.25 | * |
| dN | 0.56 | 0.32 | ** | |||
| dS | 0.04 | 0.002 | ||||
| Control | 35 | dN/dS | 0.74 | 0.55 | *** | |
| dN | 0.76 | 0.58 | *** | |||
| dS | 0.49 | 0.24 | ** | |||
| falciparum-reichenowi versus yoelii-berghei | Ligand | 21 | dN/dS | 0.46 | 0.21 | * |
| dN | 0.41 | 0.17 | ||||
| dS | −0.01 | 0.0002 | ||||
| Control | 26 | dN/dS | 0.59 | 0.35 | ** | |
| dN | 0.39 | 0.15 | * | |||
| dS | −0.19 | 0.03 | ||||
| vivax-knowlesi versus yoelii-berghei | Ligand | 25 | dN/dS | 0.56 | 0.31 | ** |
| dN | 0.54 | 0.30 | ** | |||
| dS | 0.25 | 0.06 | ||||
| Control | 67 | dN/dS | 0.66 | 0.44 | *** | |
| dN | 0.54 | 0.29 | *** | |||
| dS | 0.38 | 0.15 | ** |
N = number of gene loci analysed for the pairwise correlations between each independent species pair. * P<0.05, ** P<0.01, *** P<0.001
Figure 2. Scatterplots of dN/dS estimates for orthologous loci in independent Plasmodium species pairs.
A. vivax-knowlesi vs. falciparum-reichenowi, B. yoelii-berghei vs. falciparum-reichenowi and C. yoelii-berghei vs. vivax-knowlesi. A line of identity representing equal selective constraint and/or positive selection in orthologous genes in different species is shown on each plot (dotted line). Filled squares represent gene pairs from the control gene dataset, open squares gene pairs from the set of candidate ligand genes. Sample sizes and results of Spearman's rank correlation analysis are shown in Table 2.
Thus, although broadly similar signatures indicating positive selection on distinct classes of genes may be seen in different parts of the Plasmodium phylogeny, predictions about positive selection on individual genes for which sequence data are currently missing in particular species cannot be reliably extrapolated from orthologues in other parts of the phylogeny. To detect loci that have undergone positive directional selection in the lineage of a particular species, sequences must be directly compared with orthologues of a closely related species. As P. falciparum is currently the most important human parasite, completion of the closely related P. reichenowi genome sequence should now have particularly high priority [3].
Materials and Methods
Sets of candidate genes and controls
A set of 55 single-locus genes encoding surface proteins that are putatively ligands at various life cycle stages was first defined. These genes are candidates to display signatures of positive selection due to their likely role in host-parasite interaction, and of these, 43 could be included in comparative dN/dS analyses as noted in the following section. Loci in this candidate gene dataset were compared with loci from a control dataset chosen to represent an unbiased sample of genes not hypothesised to be under positive selection. The control set was of loci on P. falciparum chromosome 3 that contained one or more nucleotide difference among the sequences of five isolates as published [8] with data searchable on PlasmoDB (www.plasmodb.org) [9]. Of the 104 such loci identified, two (PFC0210c and PFC0420w) were already included in the candidate ligand gene dataset and were thus excluded from the control dataset, which therefore consisted of 102 genes.
Defining orthologous genes for analysis of sequence divergence between species
Pairwise nucleotide divergence was estimated for 3 pairs of closely related species: P. falciparum and P. reichenowi; P. vivax and P. knowlesi; and P. yoelii and P. berghei (Fig. 1A). Two other species for which genome sequence data are available, P. gallinaceum and P. chabaudi, were not included in the present analysis as the former is not very closely related to any other species [10]–[13], and the latter would add little extra information to the yoelii-berghei pair [7]. Protein-coding gene sequences in the P. falciparum 3D7 genome sequence (release date 11/02/2005), produced by a consortium of the Wellcome Trust Sanger Institute (WTSI), the Institute for Genomic Research (TIGR) and Stanford University [14], were downloaded from the PlasmoDB website (http://www.plasmodb.org/common/downloads/) [9]; sequences from P. vivax (release date 03/11/2005) and P. yoelii (23/07/2004) [15], produced by the Institute for Genomic Research (TIGR), were downloaded from the TIGR website (ftp://ftp.tigr.org/pub/data/Eukaryotic_Projects/); shotgun sequences from P. reichenowi (11/03/2004) [3], and gene sequences from P. knowlesi (06/01/2006) and P. berghei (08/06/2004) [7] were produced by the Wellcome Trust Sanger Institute and were downloaded from the WTSI website (ftp://ftp.sanger.ac.uk/pub/pathogens/).
Orthologues to P. falciparum predicted protein sequences were defined by BLASTp (protein vs. protein) searches against databases of P. yoelii, P. berghei, P. vivax and P. knowlesi predicted proteins, and required a reciprocal best match against the P. falciparum predicted protein database. For added stringency, each pair of putative orthologues (yoelii-berghei, vivax-knowlesi, falciparum-reichenowi) were BLASTed against the database of the other species of the pair to ensure that the best matches to the P. falciparum sequences in each species were also reciprocal best matches to each other. Where this was not the case the pair was not analysed (detailed results of BLAST searches are shown in tables S1 and S2).
No database of predicted proteins existed for P. reichenowi, so P. falciparum predicted protein sequences were used to search the P. reichenowi genomic contig database using tBLASTn (protein versus DNA translated in all 6 possible reading frames). In a number of cases where P. reichenowi orthologues could not be identified in the contig data, published P. reichenowi sequences were obtained from GenBank or sequences built from shotgun sequencing reads were used (table S3). For each gene, the P. falciparum coding sequence (introns excluded) was aligned to the best matching P. reichenowi contig using the SeqMan II program (DNASTAR, Madison, WI) to define the start and end of the coding sequence and the intron-exon boundaries. P. reichenowi contig sequences contained some regions of single-read coverage, so nucleotide mismatches in regions of single-read coverage were edited to match the P. falciparum sequence, and only well supported nucleotide mismatches in regions of multiple-read coverage were used for analysis. If a P. reichenowi gene sequence contained apparent frameshifts supported by multiple-read coverage, it was considered to be a pseudogene and not analysed.
Forty three of the 55 candidate ligand gene loci examined could be analysed for pairwise divergence between orthologues. Twelve loci (msp1, msp2, msp3, msp6, msp7, eba175, eba165, ebl1, rh1, rh2a, rh2b, rh3) were not analysed, because (i) unambiguous orthologues could not be defined, or (ii) molecular evolution appeared complex such that dN and dS may not represent the accumulation of substitutions between species (some genes had dimorphic alleles that were more divergent than the paired species sequence, and others showed evidence of gene conversion with paralogues), or (iii) an orthologue appeared to be a pseudogene. Of the 102 control gene loci, all orthologous pairs identified were analysed unless they contained a pseudogene. The relatively low number of falciparum-reichenowi gene pairs analysed (37 of 102 control genes, compared to 92 and 70 for the other species pairs) reflects the low sequence coverage of the P. reichenowi genome to date.
Analysis of pairwise between-species dN and dS values for individual genes
Orthologous protein sequence pairs were aligned using clustalW [16] and the protein alignments imposed upon the nucleotide sequences using the program pal2nal [17]. For each sequence pair, pairwise dN, dS and dN/dS indices were estimated by maximum likelihood using the codeml program [18]. Maximum likelihood estimates of dN/dS were used since they are more accurate than approximate methods such as the Nei-Gojobori method when transition/transversion rate biases and nucleotide composition or codon frequency biases exist [19]. Three independent runs of codeml were made with different initial estimates for the transition/transversion rate ratio (k) and the dN/dS ratio (run 1: k = 1, dN/dS = 1; run 2: k = 0.1, dN/dS = 10; run 3: k = 10, dN/dS = 0.1) so that each run began at a different point in the likelihood space. If different final results were obtained between runs, those with the highest log likelihood value (lnL) were used, lower lnL values being assumed to represent local likelihood optima. Non-parametric statistical tests, Wilcoxon's rank sum test and Spearman's rank correlation, were carried out using STATA 9 (StatCorp LP, Texas, USA) as the indices of divergence were not assumed to be normally distributed.
Supporting Information
Results of BLAST sequence similarity searches to determine orthologuous pairs of loci in six Plasmodium genomes for 43 candidate ligand genes
(0.08 MB XLS)
Results of BLAST sequence similarity searches to determine orthologuous pairs of loci in six Plasmodium genomes for 102 genes on P. falciparum chromosome 3.
(0.12 MB XLS)
Shotgun sequencing reads used to build 7 of the P. reichenowi gene sequences
(0.03 MB DOC)
Acknowledgments
We thank Dr Richard Emes (University College London) and colleagues at the London School of Hygiene and Tropical Medicine for helpful discussions on the analyses. We also thank the Wellcome Trust Sanger Institute (WTSI) and the Institute for Genomic Research (TIGR) for making public the data from Plasmodium genome sequencing projects.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported in part by a UK Medical Research Council PhD studentship for GDW, and Wellcome Trust programme grant 074695/Z/04/B to DJC.
References
- 1.Nielsen R. Statistical tests of selective neutrality in the age of genomics. Heredity. 2001;86:641–647. doi: 10.1046/j.1365-2540.2001.00895.x. [DOI] [PubMed] [Google Scholar]
- 2.Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 3.Jeffares DC, Pain A, Berry A, Cox AV, Stalker J, et al. Genome variation and evolution of the malaria parasite Plasmodium falciparum. Nat Genet. 2007;39:120–125. doi: 10.1038/ng1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mu J, Awadalla P, Duan J, McGee KM, Keebler J, et al. Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome. Nat Genet. 2007;39:126–130. doi: 10.1038/ng1924. [DOI] [PubMed] [Google Scholar]
- 5.Volkman SK, Sabeti PC, DeCaprio D, Neafsey DE, Schaffner SF, et al. A genome-wide map of diversity in Plasmodium falciparum. Nat Genet. 2007;39:113–119. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- 6.Hughes AL, Friedman R. Amino acid sequence constraint and gene expression pattern across the life history in the malaria parasite Plasmodium falciparum. Mol Biochem Parasitol. 2005;142:170–176. doi: 10.1016/j.molbiopara.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 7.Hall N, Karras M, Raine JD, Carlton JM, Kooij TW, et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–86. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- 8.Mu J, Duan J, Makova KD, Joy DA, Huynh CQ, et al. Chromosome-wide SNPs reveal an ancient origin for Plasmodium falciparum. Nature. 2002;418:323–326. doi: 10.1038/nature00836. [DOI] [PubMed] [Google Scholar]
- 9.Stoeckert CJ, Jr, Fischer S, Kissinger JC, Heiges M, Aurrecoechea C, et al. PlasmoDB v5: new looks, new genomes. Trends Parasitol. 2006;22:543–546. doi: 10.1016/j.pt.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Perkins SL, Schall JJ. A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J Parasitol. 2002;88:972–978. doi: 10.1645/0022-3395(2002)088[0972:AMPOMP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 11.Arisue N, Hirai M, Arai M, Matsuoka H, Horii T. Phylogeny and evolution of the SERA multigene family in the genus Plasmodium. J Mol Evol. 2007;65:82–91. doi: 10.1007/s00239-006-0253-1. [DOI] [PubMed] [Google Scholar]
- 12.Rich SM, Ayala FJ. Progress in malaria research: the case for phylogenetics. Adv Parasitol. 2003;54:255–280. doi: 10.1016/s0065-308x(03)54005-2. [DOI] [PubMed] [Google Scholar]
- 13.Escalante AA, Ayala FJ. Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. Proc Natl Acad Sci USA. 1994;91:11373–11377. doi: 10.1073/pnas.91.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardner MJ, Hall N, Fung E, White O, Berriman M, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlton JM, Angiuoli SV, Suh BB, Kooij TW, Pertea M, et al. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature. 2002;419:512–519. doi: 10.1038/nature01099. [DOI] [PubMed] [Google Scholar]
- 16.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 19.Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- 20.Marshall VM, Silva A, Foley M, Cranmer S, Wang L, et al. A second merozoite surface protein (MSP-4) of Plasmodium falciparum that contains an epidermal growth factor-like domain. Infect Immun. 1997;65:4460–4467. doi: 10.1128/iai.65.11.4460-4467.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanders PR, Kats LM, Drew DR, O'Donnell RA, O'Neill M, et al. A set of glycosylphosphatidyl inositol-anchored membrane proteins of Plasmodium falciparum is refractory to genetic deletion. Infect Immun. 2006;74:4330–4338. doi: 10.1128/IAI.00054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall VM, Tieqiao W, Coppel RL. Close linkage of three merozoite surface protein genes on chromosome 2 of Plasmodium falciparum. Mol Biochem Parasitol. 1998;94:13–25. doi: 10.1016/s0166-6851(98)00045-0. [DOI] [PubMed] [Google Scholar]
- 23.Muller HM, Reckmann I, Hollingdale MR, Bujard H, Robson KJ, et al. Thrombospondin related anonymous protein (TRAP) of Plasmodium falciparum binds specifically to sulfated glycoconjugates and to HepG2 hepatoma cells suggesting a role for this molecule in sporozoite invasion of hepatocytes. Embo J. 1993;12:2881–2889. doi: 10.1002/j.1460-2075.1993.tb05950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sultan AA, Thathy V, Frevert U, Robson KJ, Crisanti A, et al. TRAP is necessary for gliding motility and infectivity of Plasmodium sporozoites. Cell. 1997;90:511–522. doi: 10.1016/s0092-8674(00)80511-5. [DOI] [PubMed] [Google Scholar]
- 25.McCormick CJ, Tuckwell DS, Crisanti A, Humphries MJ, Hollingdale MR. Identification of heparin as a ligand for the A-domain of Plasmodium falciparum thrombospondin-related adhesion protein. Mol Biochem Parasitol. 1999;100:111–124. doi: 10.1016/s0166-6851(99)00052-3. [DOI] [PubMed] [Google Scholar]
- 26.Maier AG, Duraisingh MT, Reeder JC, Patel SS, Kazura JW, et al. Plasmodium falciparum erythrocyte invasion through glycophorin C and selection for Gerbich negativity in human populations. Nat Med. 2003;9:87–92. doi: 10.1038/nm807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narum DL, Fuhrmann SR, Luu T, Sim BK. A novel Plasmodium falciparum erythrocyte binding protein-2 (EBP2/BAEBL) involved in erythrocyte receptor binding. Mol Biochem Parasitol. 2002;119:159–168. doi: 10.1016/s0166-6851(01)00428-5. [DOI] [PubMed] [Google Scholar]
- 28.Thompson JK, Triglia T, Reed MB, Cowman AF. A novel ligand from Plasmodium falciparum that binds to a sialic acid-containing receptor on the surface of human erythrocytes. Mol Microbiol. 2001;41:47–58. doi: 10.1046/j.1365-2958.2001.02484.x. [DOI] [PubMed] [Google Scholar]
- 29.Pearce JA, Mills K, Triglia T, Cowman AF, Anders RF. Characterisation of two novel proteins from the asexual stage of Plasmodium falciparum, H101 and H103. Mol Biochem Parasitol. 2005;139:141–151. doi: 10.1016/j.molbiopara.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 30.Ghai M, Dutta S, Hall T, Freilich D, Ockenhouse CF. Identification, expression, and functional characterization of MAEBL, a sporozoite and asexual blood stage chimeric erythrocyte-binding protein of Plasmodium falciparum. Mol Biochem Parasitol. 2002;123:35–45. doi: 10.1016/s0166-6851(02)00123-8. [DOI] [PubMed] [Google Scholar]
- 31.Kariu T, Yuda M, Yano K, Chinzei Y. MAEBL is essential for malarial sporozoite infection of the mosquito salivary gland. J Exp Med. 2002;195:1317–1323. doi: 10.1084/jem.20011876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Preiser P, Renia L, Singh N, Balu B, Jarra W, et al. Antibodies against MAEBL ligand domains M1 and M2 inhibit sporozoite development in vitro. Infect Immun. 2004;72:3604–3608. doi: 10.1128/IAI.72.6.3604-3608.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilberger T-M, Thompson JK, Triglia T, Good RT, Duraisingh MT, et al. A novel erythrocyte binding antigen-175 paralogue from Plasmodium falciparum defines a new trypsin-resistant receptor on human erythrocytes. J Biol Chem. 2003;278:14480–14486. doi: 10.1074/jbc.M211446200. [DOI] [PubMed] [Google Scholar]
- 34.Howard RF, Reese RT. Plasmodium falciparum: hetero-oligomeric complexes of rhoptry polypeptides. Exp Parasitol. 1990;71:330–342. doi: 10.1016/0014-4894(90)90038-e. [DOI] [PubMed] [Google Scholar]
- 35.Ridley RG, Lahm H-W, Takacs B, Scaife JG. Genetic and structural relationships between components of a protective rhoptry antigen complex from Plasmodium falciparum. Mol Biochem Parasitol. 1991;47:245–247. doi: 10.1016/0166-6851(91)90184-8. [DOI] [PubMed] [Google Scholar]
- 36.Hisaeda H, Stowers AW, Tsuboi T, Collins WE, Sattabongkot JS, et al. Antibodies to malaria vaccine candidates Pvs25 and Pvs28 completely block the ability of Plasmodium vivax to infect mosquitoes. Infect Immun. 2000;68:6618–6623. doi: 10.1128/iai.68.12.6618-6623.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomas AM, Margos G, Dimopoulos G, van Lin LH, de Koning-Ward TF, et al. P25 and P28 proteins of the malaria ookinete surface have multiple and partially redundant functions. Embo J. 2001;20:3975–3983. doi: 10.1093/emboj/20.15.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Schaijk BC, van Dijk MR, van de Vegte-Bolmer M, van Gemert GJ, van Dooren MW, et al. Pfs47, paralog of the male fertility factor Pfs48/45, is a female specific surface protein in Plasmodium falciparum. Mol Biochem Parasitol. 2006;149:216–222. doi: 10.1016/j.molbiopara.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 39.Gaur D, Singh S, Jiang L, Diouf A, Miller LH. Recombinant Plasmodium falciparum reticulocyte homology protein 4 binds to erythrocytes and blocks invasion. Proc Natl Acad Sci U S A. 2007;104:17789–17794. doi: 10.1073/pnas.0708772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaneko O, Mu J, Tsuboi T, Su X, Torii M. Gene structure and expression of a Plasmodium falciparum 220-kDa protein homologous to the Plasmodium vivax reticulocyte binding proteins. Mol Biochem Parasitol. 2002;121:275–278. doi: 10.1016/s0166-6851(02)00042-7. [DOI] [PubMed] [Google Scholar]
- 41.Stubbs J, Simpson KM, Triglia T, Plouffe D, Tonkin CJ, et al. Molecular mechanism for switching of P. falciparum invasion pathways into human erythrocytes. Science. 2005;309:1384–1387. doi: 10.1126/science.1115257. [DOI] [PubMed] [Google Scholar]
- 42.Ishino T, Chinzei Y, Yuda M. Two proteins with 6-cys motifs are required for malarial parasites to commit to infection of the hepatocyte. Mol Microbiol. 2005;58:1264–1275. doi: 10.1111/j.1365-2958.2005.04801.x. [DOI] [PubMed] [Google Scholar]
- 43.Sterkers Y, Scheidig C, da Rocha M, Lepolard C, Gysin J, et al. Members of the low-molecular-mass rhoptry protein complex of Plasmodium falciparum bind to the surface of normal erythrocytes. J Infect Dis. 2007;196:617–621. doi: 10.1086/519685. [DOI] [PubMed] [Google Scholar]
- 44.Sanders PR, Gilson PR, Cantin GT, Greenbaum DC, Nebl T, et al. Distinct protein classes including novel merozoite surface antigens in Raft-like membranes of Plasmodium falciparum. J Biol Chem. 2005;280:40169–40176. doi: 10.1074/jbc.M509631200. [DOI] [PubMed] [Google Scholar]
- 45.Black CG, Wang L, Wu T, Coppel RL. Apical location of a novel EGF-like domain-containing protein of Plasmodium falciparum. Mol Biochem Parasitol. 2003;127:59–68. doi: 10.1016/s0166-6851(02)00308-0. [DOI] [PubMed] [Google Scholar]
- 46.Silvie O, Franetich JF, Charrin S, Mueller MS, Siau A, et al. A role for apical membrane antigen 1 during invasion of hepatocytes by Plasmodium falciparum sporozoites. J Biol Chem. 2004;279:9490–9496. doi: 10.1074/jbc.M311331200. [DOI] [PubMed] [Google Scholar]
- 47.Triglia T, Healer J, Caruana SR, Hodder AN, Anders RF, et al. Apical membrane antigen 1 plays a central role in erythrocyte invasion by Plasmodium species. Mol Microbiol. 2000;38:706–718. doi: 10.1046/j.1365-2958.2000.02175.x. [DOI] [PubMed] [Google Scholar]
- 48.Kato K, Mayer DC, Singh S, Reid M, Miller LH. Domain III of Plasmodium falciparum apical membrane antigen 1 binds to the erythrocyte membrane protein Kx. Proc Natl Acad Sci U S A. 2005;102:5552–5557. doi: 10.1073/pnas.0501594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kariu T, Ishino T, Yano K, Chinzei Y, Yuda M. CelTOS, a novel malarial protein that mediates transmission to mosquito and vertebrate hosts. Mol Microbiol. 2006;59:1369–1379. doi: 10.1111/j.1365-2958.2005.05024.x. [DOI] [PubMed] [Google Scholar]
- 50.Borre MB, Dziegiel M, Hogh B, Petersen E, Rieneck K, et al. Primary structure and localization of a conserved immunogenic Plasmodium falciparum glutamate rich protein (GLURP) expressed in both the preerythrocytic and erythrocytic stages of the vertebrate life cycle. Mol Biochem Parasitol. 1991;49:119–131. doi: 10.1016/0166-6851(91)90135-s. [DOI] [PubMed] [Google Scholar]
- 51.Trenholme KR, Gardiner DL, Holt DC, Thomas EA, Cowman AF, et al. clag9: A cytoadherence gene in Plasmodium falciparum essential for binding of parasitized erythrocytes to CD36. Proc Natl Acad Sci U S A. 2000;97:4029–4033. doi: 10.1073/pnas.040561197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ling IT, Florens L, Dluzewski AR, Kaneko O, Grainger M, et al. The Plasmodium falciparum clag9 gene encodes a rhoptry protein that is transferred to the host erythrocyte upon invasion. Mol Microbiol. 2004;52:107–118. doi: 10.1111/j.1365-2958.2003.03969.x. [DOI] [PubMed] [Google Scholar]
- 53.Yuda M, Sakaida H, Chinzei Y. Targeted disruption of the Plasmodium berghei CTRP gene reveals its essential role in malaria infection of the vector mosquito. J Exp Med. 1999;190:1711–1716. doi: 10.1084/jem.190.11.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dessens JT, Beetsma AL, Dimopoulos G, Wengelnik K, Crisanti A, et al. CTRP is essential for mosquito infection by malaria ookinetes. Embo J. 1999;18:6221–6227. doi: 10.1093/emboj/18.22.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frevert U, Sinnis P, Cerami C, Shreffler W, Takacs B, et al. Malaria circumsporozoite protein binds to heparan sulfate proteoglycans associated with the surface membrane of hepatocytes. J Exp Med. 1993;177:1287–1298. doi: 10.1084/jem.177.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cerami C, Frevert U, Sinnis P, Takacs B, Clavijo P, et al. The basolateral domain of the hepatocyte plasma membrane bears receptors for the circumsporozoite protein of Plasmodium falciparum sporozoites. Cell. 1992;70:1021–1033. doi: 10.1016/0092-8674(92)90251-7. [DOI] [PubMed] [Google Scholar]
- 57.Sidjanski SP, Vanderberg JP, Sinnis P. Anopheles stephensi salivary glands bear receptors for region I of the circumsporozoite protein of Plasmodium falciparum. Mol Biochem Parasitol. 1997;90:33–41. doi: 10.1016/s0166-6851(97)00124-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ling IT, Kaneko O, Narum DL, Tsuboi T, Howell S, et al. Characterisation of the rhoph2 gene of Plasmodium falciparum and Plasmodium yoelii. Mol Biochem Parasitol. 2003;127:47–57. doi: 10.1016/s0166-6851(02)00302-x. [DOI] [PubMed] [Google Scholar]
- 59.van Dijk MR, Janse CJ, Thompson J, Waters AP, Braks JA, et al. A central role for P48/45 in malaria parasite male gamete fertility. Cell. 2001;104:153–164. doi: 10.1016/s0092-8674(01)00199-4. [DOI] [PubMed] [Google Scholar]
- 60.Cowman AF, Baldi DL, Healer J, Mills KE, O'Donnell RA, et al. Functional analysis of proteins involved in Plasmodium falciparum merozoite invasion of red blood cells. FEBS Lett. 2000;476:84–88. doi: 10.1016/s0014-5793(00)01703-8. [DOI] [PubMed] [Google Scholar]
- 61.Lustigman S, Anders RF, Brown GV, Coppel RL. A component of an antigenic rhoptry complex of Plasmodium falciparum is modified after merozoite invasion. Mol Biochem Parasitol. 1988;30:217–224. doi: 10.1016/0166-6851(88)90090-4. [DOI] [PubMed] [Google Scholar]
- 62.Dessens JT, Mendoza J, Claudianos C, Vinetz JM, Khater E, et al. Knockout of the rodent malaria parasite chitinase pbCHT1 reduces infectivity to mosquitoes. Infect Immun. 2001;69:4041–4047. doi: 10.1128/IAI.69.6.4041-4047.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsai YL, Hayward RE, Langer RC, Fidock DA, Vinetz JM. Disruption of Plasmodium falciparum chitinase markedly impairs parasite invasion of mosquito midgut. Infect Immun. 2001;69:4048–4054. doi: 10.1128/IAI.69.6.4048-4054.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thompson J, Cooke RE, Moore S, Anderson LF, Janse CJ, et al. PTRAMP; a conserved Plasmodium thrombospondin-related apical merozoite protein. Mol Biochem Parasitol. 2004;134:225–232. doi: 10.1016/j.molbiopara.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 65.Topolska AE, Lidgett A, Truman D, Fujioka H, Coppel RL. Characterization of a membrane-associated rhoptry protein of Plasmodium falciparum. J Biol Chem. 2004;279:4648–4656. doi: 10.1074/jbc.M307859200. [DOI] [PubMed] [Google Scholar]
- 66.Eksi S, Czesny B, van Gemert GJ, Sauerwein RW, Eling W, et al. Malaria transmission-blocking antigen, Pfs230, mediates human red blood cell binding to exflagellating male parasites and oocyst production. Mol Microbiol. 2006;61:991–998. doi: 10.1111/j.1365-2958.2006.05284.x. [DOI] [PubMed] [Google Scholar]
- 67.Quakyi IA, Carter R, Rener J, Kumar N, Good MF, et al. The 230-kDa gamete surface protein of Plasmodium falciparum is also a target for transmission-blocking antibodies. J Immunol. 1987;139:4213–4217. [PubMed] [Google Scholar]
- 68.Kaneko O, Yim Lim BY, Iriko H, Ling IT, Otsuki H, et al. Apical expression of three RhopH1/Clag proteins as components of the Plasmodium falciparum RhopH complex. Mol Biochem Parasitol. 2005;143:20–28. doi: 10.1016/j.molbiopara.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 69.Chattopadhyay R, Rathore D, Fujioka H, Kumar S, de la Vega P, et al. PfSPATR, a Plasmodium falciparum protein containing an altered thrombospondin type I repeat domain is expressed at several stages of the parasite life cycle and is the target of inhibitory antibodies. J Biol Chem. 2003;278:25977–25981. doi: 10.1074/jbc.M300865200. [DOI] [PubMed] [Google Scholar]
- 70.Burns JM, Jr, Adeeku EK, Dunn PD. Protective immunization with a novel membrane protein of Plasmodium yoelii-infected erythrocytes. Infect Immun. 1999;67:675–680. doi: 10.1128/iai.67.2.675-680.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dessens JT, Siden-Kiamos I, Mendoza J, Mahairaki V, Khater E, et al. SOAP, a novel malaria ookinete protein involved in mosquito midgut invasion and oocyst development. Mol Microbiol. 2003;49:319–329. doi: 10.1046/j.1365-2958.2003.03566.x. [DOI] [PubMed] [Google Scholar]
- 72.Yuda M, Yano K, Tsuboi T, Torii M, Chinzei Y. von Willebrand Factor A domain-related protein, a novel microneme protein of the malaria ookinete highly conserved throughout Plasmodium parasites. Mol Biochem Parasitol. 2001;116:65–72. doi: 10.1016/s0166-6851(01)00304-8. [DOI] [PubMed] [Google Scholar]
- 73.Shi Q, Cernetich-Ott A, Lynch MM, Burns JM., Jr Expression, localization, and erythrocyte binding activity of Plasmodium yoelii merozoite surface protein-8. Mol Biochem Parasitol. 2006;149:231–241. doi: 10.1016/j.molbiopara.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 74.Kadota K, Ishino T, Matsuyama T, Chinzei Y, Yuda M. Essential role of membrane-attack protein in malarial transmission to mosquito host. Proc Natl Acad Sci U S A. 2004;101:16310–16315. doi: 10.1073/pnas.0406187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kushwaha A, Perween A, Mukund S, Majumdar S, Bhardwaj D, et al. Amino terminus of Plasmodium falciparum acidic basic repeat antigen interacts with the erythrocyte membrane through band 3 protein. Mol Biochem Parasitol. 2002;122:45–54. doi: 10.1016/s0166-6851(02)00077-4. [DOI] [PubMed] [Google Scholar]
- 76.Li X, Chen H, Oo TH, Daly TM, Bergman LW, et al. A co-ligand complex anchors Plasmodium falciparum merozoites to the erythrocyte invasion receptor band 3. J Biol Chem. 2004;279:5765–5771. doi: 10.1074/jbc.M308716200. [DOI] [PubMed] [Google Scholar]
- 77.Ishino T, Orito Y, Chinzei Y, Yuda M. A calcium-dependent protein kinase regulates Plasmodium ookinete access to the midgut epithelial cell. Mol Microbiol. 2006;59:1175–1184. doi: 10.1111/j.1365-2958.2005.05014.x. [DOI] [PubMed] [Google Scholar]
- 78.Siden-Kiamos I, Ecker A, Nyback S, Louis C, Sinden RE, et al. Plasmodium berghei calcium-dependent protein kinase 3 is required for ookinete gliding motility and mosquito midgut invasion. Mol Microbiol. 2006;60:1355–1363. doi: 10.1111/j.1365-2958.2006.05189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Results of BLAST sequence similarity searches to determine orthologuous pairs of loci in six Plasmodium genomes for 43 candidate ligand genes
(0.08 MB XLS)
Results of BLAST sequence similarity searches to determine orthologuous pairs of loci in six Plasmodium genomes for 102 genes on P. falciparum chromosome 3.
(0.12 MB XLS)
Shotgun sequencing reads used to build 7 of the P. reichenowi gene sequences
(0.03 MB DOC)