Abstract
Background
Fatty acids are indispensable constituents of mycolic acids that impart toughness & permeability barrier to the cell envelope of M. tuberculosis. Biotin is an essential co-factor for acetyl-CoA carboxylase (ACC) the enzyme involved in the synthesis of malonyl-CoA, a committed precursor, needed for fatty acid synthesis. Biotin carboxyl carrier protein (BCCP) provides the co-factor for catalytic activity of ACC.
Methodology/Principal Findings
BPL/BirA (Biotin Protein Ligase), and its substrate, biotin carboxyl carrier protein (BCCP) of Mycobacterium tuberculosis (Mt) were cloned and expressed in E. coli BL21. In contrast to EcBirA and PhBPL, the ∼29.5 kDa MtBPL exists as a monomer in native, biotin and bio-5′AMP liganded forms. This was confirmed by molecular weight profiling by gel filtration on Superdex S-200 and Dynamic Light Scattering (DLS). Computational docking of biotin and bio-5′AMP to MtBPL show that adenylation alters the contact residues for biotin. MtBPL forms 11 H-bonds with biotin, relative to 35 with bio-5′AMP. Docking simulations also suggest that bio-5′AMP hydrogen bonds to the conserved ‘GRGRRG’ sequence but not biotin. The enzyme catalyzed transfer of biotin to BCCP was confirmed by incorporation of radioactive biotin and by Avidin blot. The Km for BCCP was ∼5.2 µM and ∼420 nM for biotin. MtBPL has low affinity (Kb = 1.06×10−6 M) for biotin relative to EcBirA but their Km are almost comparable suggesting that while the major function of MtBPL is biotinylation of BCCP, tight binding of biotin/bio-5′AMP by EcBirA is channeled for its repressor activity.
Conclusions/Significance
These studies thus open up avenues for understanding the unique features of MtBPL and the role it plays in biotin utilization in M. tuberculosis.
Introduction
Tuberculosis still remains a major cause of death, especially in developing countries. The resurgence of this disease has primarily been due to the emergence of drug resistant tubercle, especially to the most effective drug, isoniazid (INH) (1–3). The ability of bacteria to survive inside hostile environment of host macrophage is primarily due to its complex lipid bilayer formed by mycolic acids, glycolipids, lipoproteins etc. In fact, it is estimated that lipids constitute 40% of mycobacterial dry cell weight. Thus, biosynthesis and assembly of mycolic acids and other lipids constitute potential targets for chemotherapeutic intervention for treating tuberculosis (4–6). M.tuberculosis genome contains ∼250 enzymes involved in fatty acid synthesis as compared to just about 50 in E.coli (7, 8). Fatty acid synthesis involves carboxylation of nascent chain which requires acetyl CoA carboxylase, a biotin dependent carboxylase which has three functionally dissimilar domains: (1) biotin carboxyl carrier protein (BCCP); (9) biotin carboxylase component that catalyzes Mg- ATP-dependent carboxylation of BCCP to form {COO}−- BCCP; and transcarboxylase component that catalyzes transfer of carboxyl group from {COO}−- BCCP to acetyl CoA to form malonyl CoA (9,10). ApoBCCP is biotinylated by Biotin Protein Ligase (BPL/BirA) to form holoBCCP, which then participates in carboxylation reaction. The biotinylation reaction by BPL is a two-step process:
Biotin+ATP→biotinyl-5′-AMP+PPi
Bio-5′-AMP+apocarboxylase (BCCP)→holocarboxylase (biotin- BCCP)+AMP
BPL (EC 6.3.4.15) catalyses transfer of biotin to an ε -amino group of a specific lysine residue, which is usually the 35th amino acid from C-terminal of apoBCCP and converts it to active holoBCCP which promotes fatty acid initiation and elongation (11, 12). The biotinylation reaction is highly specific, as BCCP subunit of biotin dependent carboxylase is the only substrate for BPL.
There are two variants of prokaryotic BPLs classified as:
Group I, a monofunctional enzyme lacks N-terminal HTH domain e.g. M.tuberculosis, A.aeolicus, P.horikoshii.
Group II, a bifunctional enzyme, has N-terminus HTH domain which attributes repressor function to the protein e.g. E.coli, B.subtilis, P.aeruginosa.
In Group II BPL, adenylated biotin (bio-5′-AMP), an intermediate of BPL/BirA catalyzed reaction complexes with the protein (E.coli BirA – 321 amino acids) to cooperatively repress genes of biotin biosynthetic pathway (12). Group I BPL (Mycobacterium tuberculosis BPL – 266 amino acids) lacks N-terminal DNA binding domain and hence probably does not function as a repressor (13). So, the mechanism of repression of biotin biosynthetic pathway of group I BPL and organisms harboring them is yet to be ascertained.
One of the most successful anti tuberculosis drug, isoniazid, also inhibits elongation of fatty acid. Thus, our interest was focused on biotin protein ligase as it globally determines level of fatty acid synthesis. In this study, we expressed purified and characterized M.tuberculosis BPL (MtBPL) and its substrate BCCP. Moreover, this is the first report on thermodynamic parameters of group I BPL.
Results
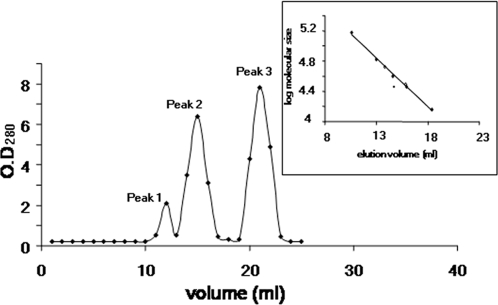
Determination of dimerization of BPL by Superdex S200 column: Equimolar concentration of purified i) BPL, ii) BPL catalysed reaction: 10 mM Tris-HCl, pH- 8.0 containing 0.1 mM BPL, 3 mM ATP, 5.5 mM MgCl2, 500 µM biotin and iii) negative control which is reaction mixture without BPL were incubated at 37°C for 1 h. The whole enzymatic process was done in the absence of BCCP, in order to determine the oligomeric status of adenylate intermediate of BPL. The reaction mixture was then individually loaded onto Superdex S-200 FPLC column. BPL without its substrate eluted as a single peak at volume corresponding to a Mr of ∼29,500±150 Da, confirming that BPL exists mostly as a monomer in its native state. This was further confirmed by Dynamic Light Scattering (DLS). MtBPL incubated with biotin also eluted as a single peak of molecular weight of ∼29,500. MtBPL catalyzed reaction with biotin and ATP resulting in bio-5′AMP, gave three peaks corresponding to a minor peak of molecular weight range Mr of 56,000±350 Da, a second peak of Mr ∼29,500±150 Da and a third peak corresponding to low molecular mass (Figure 1). The sample that eluted at lower molecular mass range (peak 3) displayed absorption in UV region and was confirmed as ATP by thin layer chromatography. The minor peak 1 corresponding to molecular weight range of Mr ∼56,000 Da was the dimeric MtBPL - bio-5′AMP - MtBPL. The second peak of molecular weight range Mr ∼29,500 was monomeric MtBPL - bio-5′AMP. Both peak 1 and 2 were holoenzyme (MtBPL - bio-5′AMP), as they biotinylated MtBCCP in the absence of biotin and ATP. To confirm if the peaks exist in equilibria, the monomeric peak was repartitioned on the same column. The monomeric species eluted as a single peak of molecular size of ∼29,500 Da. This confirms that on catalysis MtBPL forms monomeric and a weakly formed physiological dimer, both of which were holoenzymes. All the three species: apo-MtBPL, MtBPL – biotin and MtBPL - bio-5′AMP were predominantly monomers as confirmed by DLS.
Figure 1. Molecular weight determination of BPL species by gel exclusion chromatography.
BPL catalyzed reaction: 10 mM Tris-HCl p H-7.5 containing 5 nM BPL, 3 mM ATP, 5.5 mM MgCl2, 500 µM biotin but no BCCP were incubated at 37°C for 1 h. The reaction mixture was then loaded on to Superdex S 200. Peak 1: Dimeric BPL –bio 5′AMP-BPL (Mr ∼60,000±2900 Da). Peak 2: Monomeric BPL (Mr ∼29,500±150 Da). Peak 3: Unreacted ATP.
Modeling the putative amino acid residue interactions with ligand atoms:
Sequence alignment and crystal structure of Pyrococcus horikoshii BPL (PhBPL), EcBirA and MtBPL show conserved residues that are critical for substrate binding needed for enzyme activity (Table 1). The three BPLs share function and sequence homology, but differ in their oligomerization state and ligand interaction. The ligand atoms were numbered by LPC/CSU software and the ordered structure is provided in Table S1.
Table 1. Conserved residues among BPLs of different species.
| SS1 | SS2 | SS3 | SS4 | |
| PhBPL | 45GHGRLNRKW 53 | 100KWPND105 | 111K IAGVLVE118 | 124GIGLN128 |
| MtBPL | 66GRGRHGRGW 74 | 127KWPND131 | 138KLAGILAE143 | 52GVGLN156 |
| EcBirA | 115GRGRRGRKW123 | 72KWPND176 | 183 KLAGILVE190 | 203GAGIN207 |
The residues in bold are structurally similar amino acid and the residues in bold, italics are structurally unrelated amino acids.
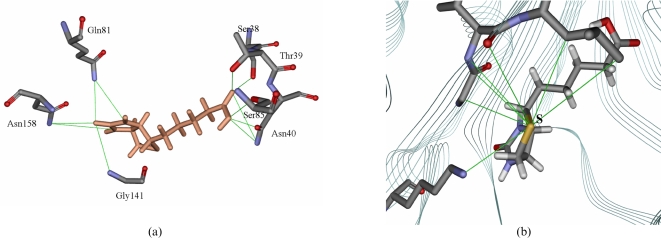
Biotin: The association of biotin and MtBPL is stabilized by 52 interactions, of which 11 are H- bonded 19 are hydrophobic interactions and 22 are other interaction viz., salt bridge and van der waal's interactions. Residues Ser38, Thr39, Asn40, Gln81, Ser85, Gly141, Asn158 form strong hydrogen bonds with biotin. The N-terminal amino acids are involved in 8 hydrogen bonds with biotin. Leu143 and Gly156 make six and two hydrophobic interactions respectively with biotin. The biotin carboxylate hydrogen bonds with side chain oxygen atoms of Ser38 and Thr39 (Figure 2a). The ureido nitrogen forms hydrogen bond with Gln81, Asn158 and the thiophene ring ‘S’ makes hydrophobic contacts with Lys138, Gly141, Ile142, Leu143 (Figure 2b;Table 2,3). The conserved ‘GXGRRG’ sequence of EcBirA and PhBPL interacts with biotin as confirmed by crystal and experimental studies. However, no such hydrogen bond interactions were observed in MtBPL - biotin association from computational analysis. The binding energy for association of biotin to MtBPL by computational analysis is – 3.645 kcal/mol.
Figure 2. Hydrogen and hydrophobic interactions between biotin and MtBPL.
(a) The H- bond interactions between biotin and the amino acid residues of MtBPL. The figure was generated using ViewerLite42. The contact of structural units (CSU) was derived with LPC software. The lines indicate H-bonds and the amino acid residues denote the interacting molecules. (b) Interaction of thiophene S atom of biotin with MtBPL. The figure was generated using ViewerLite42. The contact of structural units (CSU) was derived with LPC software. The lines indicate H-bonds and the amino acid residues denote the interacting molecules.
Table 2. List of putative hydrogen bonds between ligands biotin/bio-5′AMP and MtBPL.
| Residues | Ligands | |
| Biotin | Bio-5′AMP | |
| Ser38 Oγ | O16/3.0 | - |
| Thr39 Oγ1 | O16/3.1 | - |
| - | - | |
| Thr 39 N | - | - |
| Asn40 Oδ1 | O15/2.3 | - |
| Asn40 N δ2 | O15/3.0 | O23/4.6 |
| O35/5.3 | ||
| Asn 40 N | O16/2.9 | - |
| Gln63 Nε2 | - | O23/4.9 |
| - | O35/4.3 | |
| Ala65 O | - | O35/5.1 |
| Arg67 N | - | O23/3.8 |
| His70 O | - | N24/3.7 |
| His70 Nδ1 | - | N29/3.5 |
| - | N33/3.3 | |
| Gly71 O | - | N2/5.0 |
| - | O23/4.5 | |
| Arg72 O | - | N2/5.1 |
| Gly73 N | - | N2/4.4 |
| Trp74 O | - | N2/3.4 |
| - | N10/3.3 | |
| Trp74 | - | O13/3.7 |
| Gln81 Nε2 | O13/3.5 | - |
| N2/4.1 | - | |
| Ser85 Oγ | O15/3.4 | - |
| Asn130 Nδ.2 | - | N29/3.5 |
| - | N33/4.7 | |
| Asn130 N | - | N29/4.1 |
| Asp131 N | - | N29/4.1 |
| - | N30/4.0 | |
| Asp131 Oδ1 | - | N29/4.1 |
| - | N30/3.0 | |
| Gly141 N | O13/4.2 | O37/3.4 |
| Ile142 O | - | N26/3.7 |
| - | N30/3.5 | |
| Ile142 N | - | N26/3.8 |
| Ile142 N | - | N30/3.7 |
| Gly156 O | - | O37/3.0 |
| Asn158 Nδ2 | N10/5.3 | O36/4.4 |
| O13/3.7 | O37/4.4 | |
| Glu164 Oε1 | - | 036/5.1 |
| Glu165 Oε2 | - | N10/3.6 |
| Asp261 Oδ1 | - | N29/4.4 |
Table 3. Type of contacts made by biotin / bio-5′AMP with the MtBPL.
| Type of bonds | Biotin | Bio-5′AMP |
| Hydrogen | 11 | 35 |
| Hydrophobic | 19 | 27 |
| Acceptor-Acceptor | 3 | 4 |
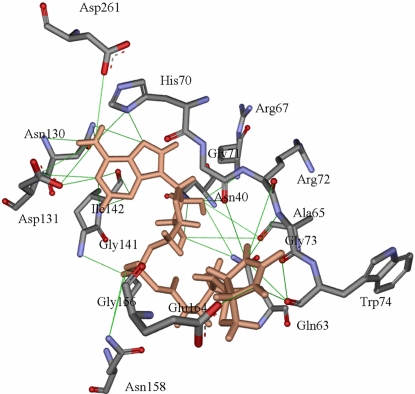
Bio-5′AMP: The association of bio-5′AMP to MtBPL is strengthened by 35 H-bonds relative to only 11 H-bonds with biotin, 27 hydrophobic interactions and 55 other weak interaction. On formation of bio-5′AMP, the interacting atoms are altered. The N-terminal amino acids Ser38, Thr39 are no longer involved in stabilization of MtBPL – bio-5′AMP complex. However, residues Gln63-Trp74 of the conserved ‘GRGRHGR’ form H-bond interaction with bio-5′AMP. Trp74 which is a conserved residue interacts with bio-5′AMP via H-bond. The ureido nitrogen interacts via H-bond with Gly71, Arg72, Gly73, and Trp74. The thiophene ‘S’ now forms strong hydrophobic interaction with Ala75, Ala76, Gln81, Ala163, Pro163. The adenine ring forms three hydrogen bonds each with His70, Asn130, Asp131, and ILe142 (Figure 3; Table 2,3). The binding energy for bio-5′AMP association to MtBPL by computational analysis is – 3.120 kcal/mol.
Figure 3. The H- bond interactions between bio-5′AMP and the amino acid residues of MtBPL.
The H- bond interactions between biotin and the amino acid residues of MtBPL. The figure was generated using ViewerLite42. The contact of structural units (CSU) was derived with LPC software. The lines indicate H-bonds and the amino acid residues denote the interacting molecules.
Desthiobiotin: Simulated docking of desthiobiotin to MtBPL show 16 H-bond and 30 hydrophobic interactions. The major difference between biotin and desthiobiotin (DTB) is the absence of thiophene ring in DTB. Hence, the contact between the thiophene ring and the protein is lacking in the analog. When the relative strengths of the bonds were analyzed, it was observed that the number of strong hydrogen bonds (donor- acceptor distance D –A<3.0 Å) were four for biotin and just one for desthiobiotin. These two factors probably contribute to the lack of desthiobiotin association with MtBPL (Figure 4b) in our experimental studies. And indeed the association of desthiobiotin to MtBPL by computational docking is unfavorable with binding energy of +0.984 kcal/mol.
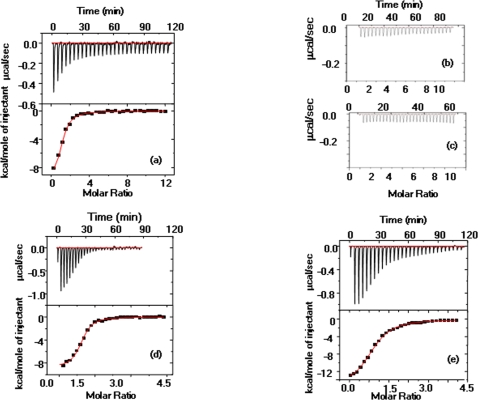
Figure 4. Isothermal calorimetry traces of the interaction of MtBPL with its ligands.
These experiments were performed in standard buffer – 10 mM Tris-HCl pH-7.5, 200 mM KCl, 2.5 mM MgCl2.Twenty nine 10 µl aliquots from a 100 µM solution of (a) biotin (b) desthiobiotin (c) ATP were used for the titrations. The lower panel shows the least squares fit of the data in the upper panel in (a) to obtain the binding parameters of the interactions of MtBPL with biotin.The fit of the data is not provided for reactions where interaction between MtBPL and a given ligand was not observed viz (b) desthiobiotin or (c) ATP. (d) MtBPL was incubated with 100 µM biotin for 30 min in the ITC cell and then titrated against twenty nine injections of 10 µl aliquots from 100 µM ATP solution. (e) Twenty nine injections of 10 µl each from a 50 µM solution of bio-5′AMP were injected into 5 µM MtBPL. The lower panel shows the least squares fit of the data in the upper panels of (d) and (e) to obtain the binding parameters of the interactions of MtBPL with biotin + ATP and Bio-5′-AMP respectively.
Isothermal calorimetry: The binding affinity of BPL for various ligands: biotin, bio-5′-AMP, desthiobiotin, ATP, ADP and AMP was determined by calorimetric titration. A typical ITC trace for binding of biotin to BPL is shown in Figure 4a. The free energy for binding of biotin to MtBPL was −8.08 kcal/mol whereas association of EcBirA-biotin is −10 kcal/mol. Biotin binds to MtBPL at a single site with high affinity of 1.06×10−6 M, a value that was less compared to that of E.coli BirA (4.2×10−8 M) but four fold greater than the Kd for biotin binding to N-terminal deleted EcBirA (65–321). MtBPL had almost similar affinities for biotin, bio-5′-AMP and biotin- ATP binary substrates. The ligands: ATP, AMP, ADP and desthiobiotin did not show any measurable enthalpy changes on association with MtBPL. Since desthiobiotin is a precursor of biotin, affinity of MtBPL for this desthio molecule was also determined. Earlier reports suggest that EcBirA can non-selectively incorporate desthiobiotin into BCCP at higher concentration. However in MtBPL, desthiobiotin association did not show appreciable heat changes even at the highest ligand concentration (1.25 mM) tested. The calorimetric titration with desthiobiotin was also performed at 2.5 fold higher enzyme concentration. Figure 4b shows the ITC trace for titration of 100 µM desthiobiotin against 10 µM MtBPL. Thus inability of MtBPL to incorporate desthiobiotin in biotinylation reaction was due to its lack of affinity for desthiobiotin.
ATP, the other substrate of MtBPL did not show appreciable heat change in calorimetric titration (Figure 4c). However, when MtBPL saturated with biotin was titrated with ATP, its dissociation constant was 1.16×10−6 M (Figure 4d). Thus, biotin binding to MtBPL opens site for binding of ATP on the protein. Bio-5′-AMP, an intermediate of enzyme catalyzed reaction was titrated against BPL. The dissociation constant for bio-5′-AMP was 1.36×10−6 M (Figure 4e), which was almost similar to that of MtBPL- biotin association. This is in contrast to that observed in EcBirA which had a 1000 fold greater affinity for bio-5′-AMP over biotin. The thermodynamic parameters governing binding of biotin to MtBPL are provided in Table 4.
Table 4. Thermodynamic parameters governing effector binding to Mycobacterium tuberculosis BPL.
| Ligands | n | Kb×106 (M−1) | ΔH (cal/mol) | ΔG (kcal/mol) | ΔS (cal/mol/K) |
| Biotin | 0.9814 | 1.06 | −11.12 | −8.087 | −10.35 |
| Biotin+ATP | 0.9229 | 1.16 | −8.18 | −9.07 | −2.925 |
| Bio-5′AMP | 0.9800 | 1.36 | −17.25 | −7.97 | −31.7 |
Titrations were carried out in 10 mM Tris-HCl, ph-7.5 containing 200 mM KCl, 2.5 mM MgCl2 using a VP-ITC calorimeter (Microcal, Inc.,USA). Binding parameters Kb and ΔH were determined b fitting of ITC data to a single-site binding model in the Origin software package. Binding free energies (ΔG) and entropies (ΔS) were calculated using the equation, ΔG = RT lnKb and ΔG = ΔH−TΔS. The values given above represent results of a single titration with each ligand and in multiple titrations the values of Kb and ΔH are reproducible within 2-fold and 1 kcal/mol.
Fluorescence titration of BPL: Figure 5 shows steady state fluorescence emission spectra of MtBPL, MtBPL + biotin. The emission maximum for MtBPL was at 342 nm similar to EcBirA. The binding of biotin to the protein resulted in 10% quenching of intrinsic fluorescence signal.
Figure 5. Flourescence titration.
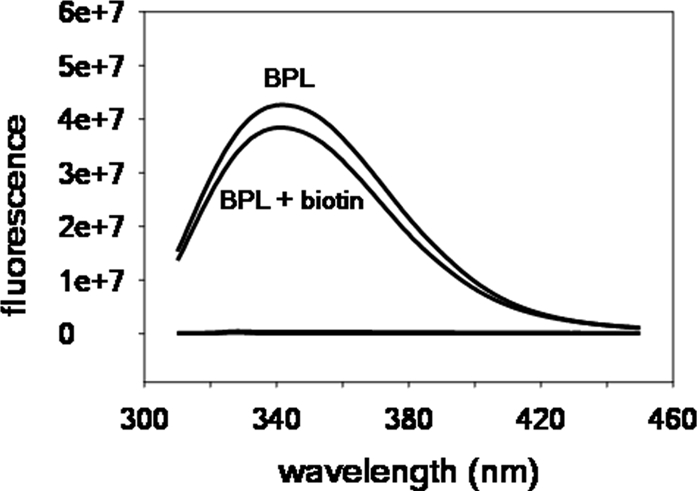
Steady state fluorescence emission spectra of MtBPL in absence and presence of biotin. [BPL] = 2 µM, [biotin] = 50 µM. All spectra were obtained in standard buffer at 20°C.
Biological properties of BPL: The enzymatic activity of BPL was determined by incorporation of 14C labelled biotin into the cloned BCCP domain of M.tuberculosis acetyl CoA carboxylase. Optimal enzyme activity was observed at pH 7.5–8.0. The presence of biotin, ATP, Magnesium ions, apo-BCCP was necessary for biotinylation reaction. Enzyme activity was dependent on ATP as nucleotide source and substitution of ATP with GTP did not result in biotinylation reaction. BPL forms a weak dimer with bio-5′-AMP synthesized from ATP and biotin, in absence of apoBCCP as observed from gel filtration (Figure 1). The Km for D-biotin is ∼424.2±56.38 nM, 21.08±3.78 µM for Mg/ATP, 5.2±.56 µM for apoBCCP and 10 nM of BPL was sufficient to drive enzymatic process (Table 5). These data thus confirm that cloned BPL of M.tuberculosis was enzymatically active.
Table 5. Kinetic constants for the interaction of Mt BPL with its substrates- biotin, MgATP and MtBCCP.
| Substrate | Km (µM) | kcat (s−1) | kcat/Km (s−1M−1)×103 |
| Biotin | 0.420±.056 | 0.034±.007 | 82±2.1 |
| Mg/ATP | 21.08±3.78 | 0.0282±.003 | 1.33±.09 |
| BCCP | 5.2±0.56 | 0.030±.001 | 5.77±.37 |
The experiment was carried out in triplicates as described in Experimental procedures.
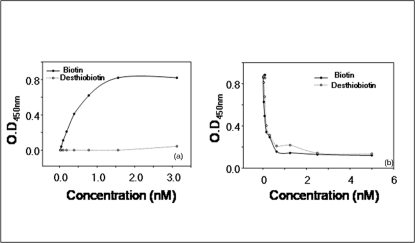
Specific incorporation of biotin by MtBPL: To determine if BPL can non-specifically utilize desthiobiotin in the enzyme reaction, both biotin and its analogue were used at concentrations ranging from 25 µM – 6 nM in biotinylation of BCCP (15 µM) by 10 nM of BPL. As revealed by an assay akin to ELISA, biotin at ∼620 nM efficiently biotinylated BCCP whereas desthiobiotin was not incorporated into BCCP in a 30 min biotinylation reaction (Figure 6a). The desthiobiotin concentration was increased to 1000 µM which was >1200 fold the optimal biotin concentration and the incubation period for 4 h. But the incorporation of desthiobiotin into BCCP by BPL was not appreciable. This could be due to two reasons: i) desthiobiotin was not utilized as a substrate in enzymatic process by BPL or, ii) streptavidin HRP was unable to detect incorporated desthiobiotin. To rule out the second possibility, a competitive assay of streptavidin with biotin/desthiobiotin was performed. Streptavidin HRP at 1∶2500 dilution of 1 mg/ml solution was pre-incubated with biotin/ desthiobiotin (5 µM–5 nM) for 1 h at 37°C and the solution was then added to previously immobilized biotinylated BCCP. Streptavidin interacts with its cognate molecule (biotin/desthiobiotin) and any unreacted, free streptavidin from first step will react with biotinylated BCCP determining affinity of streptavidin for biotin/desthiobiotin. Figure 6b shows that streptavidin interacts efficiently with both biotin and desthiobiotin. These experiments thus rule out the possibility that discrepancy in incorporation of biotin/desthiobiotin in BCCP was not due to error in detection of desthiobiotinylated BCCP but because of its non-utilization.
Figure 6. Enzyme linked immunosorbent assay to determine the substrate specificity of MtBPL.
(a) Incorporation of biotin / desthiobiotin into BCCP in BPL catalyzed reaction. The wells were coated with 15 µM BCCP and incubated overnight at 4°C. The non-specific sites were blocked with 1% BSA and biotinylation reaction carried out with biotin/desthiobiotin as the substrate. The incorporated substrate was detected streptavidin-HRP. All experiments were done in triplicates and on three different occasions. (b) Competitive ELISA to determine the affinity of streptavidin for biotin and desthiobiotin. The wells were immobilized with biotinylated BCCP. Streptavidin HRP (1∶2500) was incubated with equal volume and different concentration of biotin or desthiobiotin for 1 h at 37°C. The reaction mixture was then added to biotinylated BCCP. The unreacted streptavidin bound to immobilized BCCP which was detected by TMB/H2O2. All experiments were done in triplicates and at three different occasions.
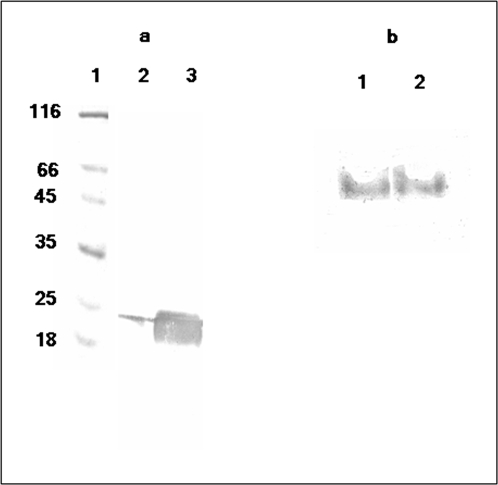
Avidin blot: Biotinylation of apo-BCCP, a biotin acceptor protein was detected by streptavidin HRP on Avidin blot (Figure 7a). Lane 2 is BCCP alone with no BPL. The faint band observed corresponds to BCCP that was probably biotinylated by endogenous EcBirA. Lane 3 is biotinylation reaction of BCCP by BPL in presence of ATP, biotin and MgCl2.for 30 min at 37°C. As observed BCCP was biotinylated several fold by BPL in comparison with BCCP control (Lane 3), thus confirming transfer of biotin to acceptor domain of BCCP by M.tuberculosis BPL. Figure 7b shows the ability of both monomeric and dimeric MtBPL associated with bio-5′AMP to biotinylate apoBCCP in the absence of biotin and ATP. The second step of the biotinylation reaction, the transfer of biotin to BCCP from bio-5′AMP by MtBPL was detected by Avidin blot (Figure 7b). Lane 1 and 2 is the peak 1 and 2 (Figure 1) of holoenzyme eluted from gel exclusion chromatography biotinylating apoBCCP.
Figure 7. Avidin blot of biotinylated BCCP catalysed by BPL.
(a) Biotinylation by BPL was carried in a reaction mixture containing 50 mM Tris pH-8.0, 3 mM ATP, 5 µM biotin, 5.5 mM MgCl2, 0.1 mM dithiothreitol, 0.1 mg/ml bovine serum albumin and 5 nM BPL and 15 µM BCCP over night at 4°C. The reaction mixture was then resolved on a 12% SDS PAGE and transferred to PVDF membrane. The membrane was then incubated with streptavidin HRP for 1 h at room temperature and developed with AEC/ H2O2. Lane 1: Molecular weight marker. Lane 2: MtBCCP was incubated with biotin, ATP and MgCl2 but no MtBPL. The band observed was due to biotinylation of BCCP by endogenous host BPL. Lane 3: MtBPL catalysed biotinylation of BCCP. The intensity of the band was several folds higher than that of biotinylation by endogenous host BPL. (b) holoMtBPL catalyzed reaction was performed in the absence of biotin and ATP. MtBCCP was incubated with sample (peak 1, 2) independently in the absence of biotin and ATP for 30 min at 37°C and resolved on a 12% SDS-PAGE and transferred to PVDF membrane. Holo-MtBPL (MtbPL-bio-5′AMP) transfers biotin to BCCP which was detected by streptavidin –HRP. Lane 1: Peak 1, Superdex S200. Lane 2: Peak 2, Superdex S200.
Discussion
Acetyl CoA carboxylase and pyruvate dehydrogenase are enzymes crucial for fatty acid biosynthesis which require biotinylation of their BCCP domain as a prelude to their enzymatic activities (14, 15). Biotin protein ligase which globally controls this pathway is a potential target for anti-mycobacterial drugs. MtBPL shares ∼25% and 31% sequence homology with PhBPL and EcBirA respectively. It belongs to monofunctional group I BPL as it lacks the 60 amino acid N-terminal HTH domain. This domain in EcBirA contributes to repressor function by binding to biotin operon (bioO). Deletion of the N-terminal domain converted EcBirA from bifunctional to monofunctional protein (16).
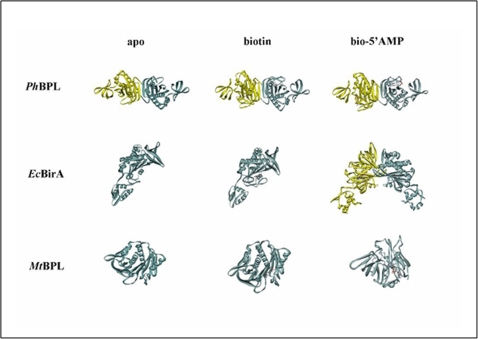
The present study shows that MtBPL differs from biotin ligase of other species both in interaction with its substrates and molecular mechanism of catalysis. Homodimerization is coupled to co- repressor synthesis and is a pre-requisite for repressor function in EcBirA (16). Monofunctional PhBPL (Group I) is a dimer in all the three forms viz. apo, biotin and bio-5′AMP liganded forms (Figure 8). PhBPL in spite of lacking repressor function does exist as a dimer. Our studies reveal that apo, biotin and bio-5′AMP liganded. MtBPL are monomers (Figure 1, 8). The peak corresponding to monomeric MtBPL (peak 2, Figure 1) was holoenzyme as it biotinylated MtBCCP. However, bio-5′AMP binding does promote weak physiological dimerization of MtBPL similar to that observed when biotin binds to EcBirA. But the ability of both peak 1 and 2 to transfer biotin to MtBCCP confirmed that both the peaks were holoenzymes. Truncated EcBirA lacks repressor function but is enzymatically active (bio-5′AMP synthesis). It would be interesting to study the oligomerization property of this truncated protein on bio-5′AMP synthesis Thus, our gel exclusion profiling studies suggest that monofunctional MtBPL does not require dimerization for catalysis. Hence from studies on different BPLs, it is evident that oligomerization may not be a universal phenomena for all BPLs and some of them such as PhBPL and EcBirA, undergo oligomerization for compaction and thermo-stability in their apo/holo forms (17, 18).
Figure 8. A schematic illustration of apo form, biotin and bio-5′AMP forms of PhBPL, MtBPL and EcBirA.
It is known that PhBPL is a dimer in all the three forms, apo, biotin and bio-5′AMP liganded, while EcBirA is a dimer when associated with bio-5′AMP but monomeric in apo and biotin-liganded form. From our studies it was observed that MtBPL is monomeric in all the three forms (Figure 8).
Sequence analysis shows conserved sequences between MtBPL, EcBirA and PhBPL. Four homologous sequences between the three BPLs are shown (19). However, the identity of other residues dispersed in the protein sequences are also conserved but are not specified. Sequence SS1 interacts with biotin in PhBPL and EcBirA. Earlier studies by Kwon and Beckett (2000) show that the consensus ‘GRGRXGR’ sequence is involved in binding of biotin and bio-5′AMP (20). Docking studies of MtBPL and biotin show that sequence SS1 was involved in interaction with bio-5′AMP but not biotin. In fact, except for residues 66Gly and 67Arg which were involved in hydrophobic interaction with the thiophene ring, the reminder of the conserved sequence appears to be of little or no consequence for the recognition of biotin. From ITC studies, it was observed that MtBPL has a dissociation constant of 1×106 M−1 for biotin (Figure 4a). Hence, the hydrogen bond between the hydrophilic residues, Ser 38, Thr39, Asn40, Gln81, Ser85 and biotin contribute to this association. The hydrophobic pocket formed by Ile83, Leu84, Ala140, Gly141, Ile142, Leu143, Gly154, Val 155, and Gly156 probably dock the thiophene ring of biotin. Bio-5′AMP interacts with Arg67, His70, Gly71, Arg72 of the conserved ‘GRGRHGR’ sequence by H-bonds and hydrophobic interaction. Asn158 contributes two H-bonds to the adenylate and the purine ring stacks on the face of the indole ring of Trp74 and makes H-bond with Asn158, and side-chain of alanine 65. An analogous interaction in EcBirA, is provided by Phe which replaces alanine of MtBPL at the homologous position. In EcBirA, biotin binding site is a hydrophobic groove with hydrogen binding partners Ser89 and Thr90 pre- positioned to accept biotin (21). From our computational studies, it was observed that homologous residues Ser38 and Thr39 of MtBPL H-bond with biotin. Thus, residues Ser38, Thr39 for biotin and ‘GRGRHGRGW’ sequence for bio-5′AMP may contribute largely to association of ligands. From docking simulations, biotin has 52 and bio-5′AMP has 117 interactions with MtBPL. The number of interactions involved and the larger surface area occupied by bio-5′AMP protects EcBirA from proteolysis. This suggests that though bio-5′AMP is an intermediate molecule; its complex with BPL is most stable. However, these computational analysis need to be confirmed by experimental studies.
The other focus of our study is the substrate specificity of MtBPL for biotin. Desthiobiotin did not bind to MtBPL nor was used in the biotinylation reaction. The thiophene ring and the hydrophobic tail would interact with the hydrophobic amino acids and the biotin carboxyl and ureido ring would interact with the hydrophilic amino acids. Since DTB lacks the thiophene ring, the interaction of the molecule with the hydrophobic amino acids is only via the hydrophobic tail and ureido ring. Also, biotin forms relatively stronger H-bonds (bond length >3.0Å) as compared to desthiobiotin. This is reflected in the unfavorable binding energy of association between desthiobiotin and MtBPL (Biotin = −3.645 kcal/mol, Desthiobiotin = +0.984 l/mol).
A second conserved sequence of KWPNDVL shares sequence homology with biotin binding protein avidin. However, this sequence does not interact with the ligands in all the three BPLs. Lys111 (PhBPL) and Lys183 (EcBirA) of the conserved ‘KXAGXLVE’ play a critical role in adenylation reaction. However, from computational studies, no such interaction between this conserved lysine residue and the ligands were observed (21, 22).
Effector binding is central to a number of essential activation processes and provides essential information on interactions involved therein. The affinity of MtBPL for its effectors biotin and bio-5′-AMP was determined by ITC. The thermodynamic data are categorized into enthalpy, free energy changes and dissociation constants. MtBPL exhibited similar dissociation constants of ∼1×10−6 M for both the effectors (Figure 4a, d and e). In contrast, EcBirA displayed different affinities for its ligands: biotin and bio-5′-AMP. Dissociation constants for biotin (4.2×10−8 M) and bio-5′AMP (4×10−11 M) was very different and hence they have been classified as ‘weak’ and ‘strong’ effectors respectively (23–25). The affinity of MtBPL for bio-5′-AMP was 5 orders of magnitude lower than that observed in EcBirA (4×10−11 M). Repressor function in EcBirA is in association with bio-5′-AMP (co-repressor) and hence very tight binding is required to perform this function (26). However, the N-terminal deleted EcBirA had 1000 fold reduced affinity (∼4×10−8 M) for bio-5′-AMP (16). This suggests that mono-functional group I BPLs exhibit attenuated affinities dedicated to perform only enzymatic function. The adenosine molecules, ATP, ADP and AMP did not show any appreciable heat changes in ITC. This is in contrast to crystallographic structure of PhBPL which show interaction with adenosine molecules (22). However, in crystallographic structures, observations of the complexes of ligand which have extremely low affinities (<50 M−1) can often be observed which is typically difficult in biophysical studies. The endoplasmic reticulum chaperone, calreticulin, for example, does not show binding to glucose, though density for glucose complexed to calnexin, a very closely related molecule was seen in the crystal structure (27).
Bio-5′AMP, an intermediate of MtBPL catalyzed reaction involves biotin and ATP as substrates. Hence, it was essential to determine the mechanism of association of MtBPL with its substrates. These studies show sequential binding pattern of MtBPL to them, as ATP (Figure 4c) does not interact with BPL directly but on saturation with biotin, it exhibits a Kd of ∼1.16×106 M (Figure 4d). Saccharomyces cerevisiae BPL interacts first with ATP followed by biotin, in contrast to MtBPL and EcBirA (28). Biotin is the limiting substrate in MtBPL catalyzed reaction similar to EcBirA. Biotin binding perhaps contributes to structural transitions that may be required for interaction of ATP to form ternary complex between MtBPL, biotin and ATP.
The Gibbs free energy for binding of biotin and bio-5′-AMP to MtBPL were almost similar. But for EcBirA, there was a gain of −4 kcal/mol of free energy on bio-5′-AMP binding. This is probably to ensure that repressor was always in active form for binding to bioO (29, 30). MtBPL does not require any enhanced BPL-bio-5′-AMP stability for transfer of biotin to apoBCCP. Thus, thermodynamic studies reveal that molecular mechanisms of ligand interaction are very different between MtBPL and EcBirA on account of additional burden of repressor function in the latter molecule.
Binding of bio-5′-AMP to MtBPL is associated with larger enthalpy changes (−17.2 kcal/mol) in comparison with −11.12 kcal/mol for biotin (Table 4). However in EcBirA, enthalpy changes are larger for biotin (−19.2 kcal/mol) compared to that for bio-5′-AMP (-12.3 kcal/mol). According to Brown and Beckett (24), the absence of a charged functional group to interact with the adenosine moiety could be the reason for the enthalpic penalty. In EcBirA, there is a biotin binding loop (BBL) 115–120 residues which is conserved and adenylate binding loop (ABL) consisting of residues 212–223 (24, 25). On limited proteolysis, the region between amino acid residues 217–218 was protected by bio-5′-AMP (26). This suggests that adenosine of bio-5′-AMP interacts at the ABL. BBL becomes ordered on biotin binding and then adenylate binding site is formed in EcBirA. In contrast, there is no separate ABL and BBL in PhBPL hence the biotin moiety of biotinyl-5′-AMP and biotin share the same binding site. Also, the adenosyl moiety of bio-5′-AMP and ADP interact almost at the same position. This sharing of binding site in PhBPL which is strikingly different from that observed in EcBirA suggests that catalytic binding sites are organized such that major conformational changes are perhaps not required in group I BPLs during the reaction (22). Bagautdinov et al (22) co-crystallized PhBPL with biotin and bio-5′AMP suggesting that the interaction does not involve any conformational change. On the other hand, exposure of EcBirA crystals to bio-5′-AMP resulted in cracking and crystal destruction due to conformational changes. Together, these data underscore striking differences in the organization of biotin and ATP binding site between MtBPL and EcBirA as well as the differences in the conformational changes they undergo upon binding of biotin/bio-5′-AMP.
The dissociation constant of MtBPL for its ligands was high relative to EcBirA (Table 4, Figure 4), hence enzyme turnover was determined by steady state kinetics. The kinetic constants show low Km for both biotin (0.42 µM) and ATP (21 µM). This is similar to that observed in eukaryotic biotin auxotrophs such as Saccharomyces cerevisiae and A.thaliana which exhibit Km in nanomolar range for biotin and lower micromolar concentration for MgATP (28, 32). EcBirA has comparable Km of 0.3 µM for biotin, but has higher Km of 300 µM for ATP. Though MtBPL has higher dissociation constant for both biotin and bio-5′-AMP, the Km values were comparable to that of EcBirA (Table 4,5). Thus, the lower binding affinity of MtBPL for its ligands did not alter its enzymatic function.
Desthiobiotin (DTB) is precursor molecule for biotin but differs from it in several ways; both have an ureido ring but desthiobiotin lacks thiophene ring (Table S1). Wu et al (33) had suggested that EcBirA incorporates desthiobiotin into SAPK, a protein with a biotinable lysine residue. Their assay was driven to 100% incorporation at a concentration of 1 mM in 4 h. But MtBPL did not promote desthiobiotin incorporation into BCCP at increased concentration and incubation time (Figure 6a,b). From structural analysis of PhBPL, it is evident that both thiophene and ureido side chains are accommodated in the hydrophilic bottom provided by glycine residues (Gly45,47,127,129) and the thiophene moiety interacts with the hydrophobic wall (22). DTB which lacks the thiophene side chain will not fit efficiently in such a structural frame work. The hexanoic acid attached close to ureido nitrogen may hamper interaction of ureido side chain as well. This argument is strengthened by the binding energy of MtBPL (+0.984 kcal/mol) and EcBirA (−1.897 kcal/mol) for desthiobiotin by computational modeling. Monofunctional MtBPL has an unfavorable binding energy for the association and this is confirmed by the inability of the enzyme to either bind or incorporate desthiobiotin. EcBirA prefers biotin (−3.491 kcal/ mol) to desthiobiotin (−1.897 kcal/mol) and hence utilizes desthiobiotin at higher molar concentration and increased incubation period. Biotin synthase (BioB) utilizes desthiobiotin to synthesize biotin and activity of the enzyme is determined by bioassay. This is a very tedious assay and there is a need for more reliable, easy, quantitative and rapid assay. Biotin synthase enzymatically generates one molecule of biotin from desthiobiotin. At equimolar concentration of biotin/desthiobiotin, MtBPL preferably incorporates biotin and this can be exploited to assay biotin synthase activity in a sensitive manner.
In summary, we have successfully cloned an enzymatically active MtBPL and biotin acceptor molecule, BCCP of M. tuberculosis. Our protocol variation of reduced growth and induction time yielded nearly 90% apo-BCCP. Our studies suggest that the mechanism of enzymatic process is at considerable variance from Group II BPL. MtBPL binds initially to biotin with a moderate affinity as compared to EcBirA. This complex then interacts with ATP to synthesize intermediate molecule bio-5′-AMP and biotin is transferred efficiently to BCCP. From our studies, it is clear that reduced affinity of MtBPL does not alter enzyme kinetic parameters, suggesting that enhanced binding affinity in EcBirA probably contributes to repressor activity. However, in this study we report that MtBPL is merely not an equivalent of N-terminal deleted EcBirA, but has its own unique modus operandi, including specificity and affinity for its ligands to promote biotinylation of BCCP domain of acetyl CoA carboxylase, an enzyme critical for survival of the pathogen. Also, the differences in the architecture of biotin and ATP binding site of MtBPL and EcBirA and the exquisite specificity of MtBPL for biotin over desthiobiotin can be harnessed for synthesizing inhibitors for this important enzyme which should aid in combating global spread of tuberculosis.
Materials and Methods
Biochemicals
D-biotin, desthiobiotin, IPTG and ATP (disodium salt) were purchased from Sigma, USA. Restriction enzymes were obtained from MBI Fermentas, USA. Ni-NTA agarose was purchased from Novagen. All buffers were prepared from reagents of standard grade.
Construction of plasmids and strains
Expression and purification of MtBPL: The BPL gene was amplified from M.tuberculosis chromosomal DNA by primers designed with restriction sites NdeI/HindIII. The PCR primers used for amplification are MtBPL F1: 5′- GGAATTCCATATGGTGACCGACCGCGAT -3′; MtBPL B1: 5′-CCCAAGCTTTTAACGCAAATGCAC-3′. The putative clone of BPL was sequenced using T7 promoter and terminator. It was cloned into pET 28a expression vector for recombinant expression in E.coli BL21 with hexahistidine tag at N-terminus. For optimal yield of recombinant protein, cells were grown at 37°C to an O.D600 ∼ 0.7 and induced with 0.1 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) for 4 h at 25°C. The cells were suspended in binding buffer (10 mM Tris-HCl, 500 mM NaCl, pH- 8.0), sonicated twice for 1 min on ice. The cell free sonicate was loaded onto charged Ni- NTA affinity resin (Novagen) and eluted with 100 mM imidazole. The eluted protein was dialyzed against standard buffer (10 mM Tris, 200 mM KCl, 2.5 mM MgCl2, pH-8.0). MtBPL was a monomer (Mr ∼29,500) in native form as determined by gel exclusion chromatography (Superdex S200) on fast pressure liquid chromatography and Dynamic Light Scattering. The purity of the protein thus purified was in the range of 95–98% as determined by SDS-PAGE.
Design of primers, expression and purification of BCCP: The BCCP of M.tuberculosis has not been annotated as an individual gene but was part of acetyl CoA carboxylase in NCBI database. Thus BCCP of M.tuberculosis was cloned on the basis of the similarity to E.coli BCCP which was annotated as a protein of 156 amino acids with lysine residue at the 35th position from C-terminal. The BCCP gene was cloned as a 468 bp PCR product amplified from M.tuberculosis chromosomal DNA as template using the primers MtBCCP F1: 5′-CGCGGATCCATGCGCGGCGGGGGCCATATG-3′ and MtBCCPF1: 5′CGCTCGAGCTAGTCCTTGATCCTCGCCAGTACC-3′.This PCR product was cloned at BamHI/XhoI sites of pET28a and the protein served as substrate for BPL. The cloned BCCP was sequence verified with T7 promoter and terminator using sequencer DNA star, Wisconsin. For optimal protein expression, cells were grown overnight in LB containing 30 µg/ml of kanamycin at 37°C for 14 h. Next day, 15% inoculum of this culture was added to fresh LB and grown for 1 h at 37°C. The culture was induced with 0.1 mM IPTG for 1 h at 25°C to prevent endogenous biotinylation by host BPL. The soluble protein did not bind to Ni- NTA agarose, probably because hexahistidine tag was masked. Hence, cells were purified under denaturation conditions (10 mM Tris, 100 mM NaH2PO4, 10 mM imidazole, 8 M Urea, pH- 8.0) and sonicated for 2 min, twice on ice. The cleared sonicate was loaded onto to a charged NiNTA column and the protein was eluted with elution buffer (10 mM Tris, 100 mM NaH2PO4, 10 mM imidazole, 8 M urea, pH- 4.5).The protein was further dialyzed against 10 mM Tris-HCl, pH- 7.5 containing decreasing concentration of urea.
Determination of protein concentration
Concentration of protein was determined by Lowry's method and ultraviolet absorbance using an extinction coefficient of 1.26 mL mg−1 cm−1 for BPL and 1.373 mL mg−1cm−1 for BCCP. Both BPL and BCCP were exhaustively dialyzed against standard buffer of 10 mM Tris-HCl (pH- 7.5), containing 200 mM KCl and 2.5 mM MgCl2.
Synthesis of Bio-5′-AMP
Bio-5′- AMP was synthesized by a modified method of Lane et al (34): To biotin (244 mg) dissolved in 4 ml of 75% aqueous pyridine with gentle heating 347 mg of ATP in 3 ml of 75% aqueous pyridine was added. To the reaction mixture, pre cooled 4.5 g of dicyclohexyl carbodiimide in 5 ml of absolute pyridine was added and reaction was carried out for 8 h at 0°C with constant stirring. The reaction mixture was kept aside at 4°C for 24 h to precipitate DCU which was removed by passing through a sintered glass filter. Bio-5′-AMP was separated from unreacted biotin and ATP by RP-HPLC performed on Waters equipment consisting of a Waters 2487 Dual Absorbance Detector. Analyses were carried out on an analytical column (Nucleosil 100 Å 5 µm C18 particles, 250×4.6 mm) using following solvent system: solvent A, 0.1% TFA in water; solvent B, 0.1% TFA in acetonitrile; flow rate, 1.0 mL min−1 with UV monitoring at 214 nm and 254 nm. Preparative column (Delta-PakTM 300Å 15 µm C18 particles, 200×2.5 mm) was used with an identical solvent system at a flow rate of 25 ml min−1. Mass spectra were obtained by electron spray ionization (ESI-MS).
Analytical gel filtration chromatography
The oligomeric status of purified BPL was determined by gel filtration on Superdex S200 column and a fast protein liquid chromatography system (Pharmacia Biotech Inc). The column dimension was 30 cm×2.3 cm. The mobile phase was 50 mM Tris-HCl buffer, pH- 7.5, and flow rate was 0.2 ml/min. The molecular mass of standard proteins in kDa 150, 66, 45, 29 & 14 were, respectively, alcohol dehydrogenase, BSA, ovalbumin, carbonic anhydrase and lysozyme. The eluted proteins were detected by their absorbance at 280 nm.
The enzymatic reaction was carried out with 100 µM BPL, 3 mM ATP and 500 µM biotin, 5.5 mM MgCl2 at 37°C for 30 min and 300 µl of reaction mixture was loaded onto the column. The void volume (Vo) was determined by calibrating column with blue dextran. From elution volume of standard proteins, molecular size of unknown protein, BPL and its enzymatic reaction mixture were determined.
Computational modeling of ligand docking to MtBPL: The structure of MtBPL was modeled using SWISS-PROT software with Mycobacterium tuberculosis Biotin Protein Ligase as the template (Code: 2cghB) as the template (Ma Q. and Wilmanns M. unpublished). The ligands: biotin, desthiobiotin and bio-5′AMP were built using MOE (Molecular Operating Environment) and energy minimized under the MMFF94×forcefield. The docking of the ligands to MtBPL was also performed with MOE using AMBER99 forcefield and the docked complex with least energy was chosen. The H bond interaction between the ligand atoms and amino acid residues of MtBPL was obtained by LPC/CSU software (35).
Assay of BPL: BPL activity was assayed by measuring incorporation of [14C]- biotin (GE Healthcare, UK) into BCCP 156 as described by Chapman-Smith et al (10,11). The reaction contained 10 mM Tris-HCl, 200 mM KCl, 3 mM ATP, 5.5 mM MgCl2, 10 µM biotin and 1 µM of [14C] biotin (sp.activity 58.0 mCi/mmol), 0.1 mM DTT, 0.1 mg bovine serum albumin and 15 µM BCCP. The reaction was initiated by addition of 10 nM of BPL and incubated at 37°C for 30 min. The reaction mixture was added to biotin and trichloroacetic acid treated filters. After air drying, the filters were washed twice with 10% ice-cold TCA, ethanol and air dried and acid insoluble radioactivity was measured. A unit of BPL activity was defined as amount of enzyme required to incorporate 1 nM of biotin per min. Values for Km and Vmax were determined by non-linear least squares fit of the plots of substrate concentration versus velocity of reaction to Michaelis-Menten equation.
Determination of thermodynamic parameters by Isothermal calorimetry
A MicroCal VP-ITC was used to determine binding parameters of biotin and desthiobiotin to BPL. All protein samples were dialyzed exhaustively against standard buffer, filtered and degassed before titration. Each calorimetric experiment was performed at 20°C; 10 µM of enzyme in reaction cell was titrated against 100 µM biotin, desthiobiotin, ATP, ADP, AMP from the syringe. But for determination of binding constant for bio-5′-AMP, 5 µM BPL was used. All heats of binding were corrected for heat of dilution by subtraction of average heat associated with multiple injections of ligand performed after saturation of protein (12). The binding constant (Kb), enthalpy (ΔH), and stoichiometry (n) of the reaction were obtained from this corrected data by non-linear least square analysis using Origin 7.0 software package. The Gibbs free energy (ΔG), and entropy (ΔS), were calculated using the equation
| (1) |
| (2) |
To determine association of biotin in presence of ATP, 10 µM of enzyme in standard buffer was allowed to equilibrate with 100 µM of biotin for 30 min in the reaction cell prior to titration with 100 µM of ATP.
Fluorescence titration
Fluorescence titrations were made using a Fluorolog, tau-3 life-time spectrofluorimeter system according to the method of Xu et al (25). Measurements were performed in buffer containing 10 mM Tris-HCl, 200 mM KCl, 2.5 mM MgCl2, pH-7.5±0.02 at 20°C.The excitation wavelength was 295 nm, and emission was monitored from 310–450 nm. The excitation and emission slit widths were maintained at 4 nm. All measurements were made in ratio mode using rhodamine B as a standard. Titrations were performed by sequential addition of ligand to enzyme. The mixture was allowed to equilibrate for 2 min prior to measurement of spectra. The spectra were corrected for sample dilution and no correction was required for inner filter effect because absorbance of ATP at 295 nm at concentrations used was less than 0.005.
Enzyme linked immunosorbent assay to determine specific incorporation of biotin
To determine preferential utilization of substrate by BPL, a method akin to ELISA was used. The maxisorp plates (Nunc) were coated overnight with 15 µM BCCP in phosphate buffered saline (PBS) at 4°C.The wells were washed with PBS and dried off buffer. The BCCP was biotinylated at 37°C for 30 min and then streptavidin conjugated horseradish peroxidase (HRP) was incubated at 1∶5000 dilutions for 1 h at 37°C. The wells were washed and bound streptavidin was detected by TMB/H2O2 (Bangalore Genei, India).
Competitive ELISA was performed to determine difference in interaction specificity of streptavidin for biotin/desthiobiotin using the ability of streptavidin to recognize biotin and desthiobiotin. In this assay, 100 µl of streptavidn HRP (1∶2500) was incubated with equal volume of varying concentration of biotin/desthiobiotin (5 µM–5 nM) for 1 h at 37°C. Then 100 µl of this solution was added to solid-phase biotinylated BCCP and incubated for 1 h at 37°C. Free unreacted streptavidin reacted with biotinylated BCCP. The bound streptavidin HRP was then detected by TMB/ H2O2.
Avidin blot: Biotinylation of BCCP by BPL was confirmed quantitatively by avidin blot. The reaction was carried out in a mixture containing 50 mM Tris, pH- 8.0, 3 mM ATP, 500 µM biotin, 5.5 mM MgCl2, 0.1 mM dithiothreitol, 0.1 mg/ml bovine serum albumin,15 µM BCCP. Reaction was initiated by 20 nM BPL at 37°C for 1 h. The reaction mixture was then resolved on 12% polyacrylamide gel and transferred to PVDF membrane. The non-specific sites on membrane were blocked with 1% BSA and then incubated with streptavidin conjugated to horseradish peroxidase (1∶2000) for 1 h at room temperature. The membrane was developed with AEC/ H2O2
Supporting Information
Structures of biotin, bio-5′AMP and desthiobiotin. The ligand atoms are numbered as per the LPC/CSU software.
(0.05 MB DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by Centre of excellence grant from the Department of Biotechnology (DBT), Government of India to A.S. and by another DBT grant to A.S. A.S. is also J.C. Bose Fellow of the Department of Science and Technology, Government of India.
References
- 1.Barry BR, Murray JL. Tuberculosis: Commentary on a re-emergent killer. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 2.Johnsson K, King DS, Schultz PG. Studies on the mechanism of action of isoniazid and ethionamide in the chemotherapy of tuberculosis. J. Am. Chem. Soc. 1995;117:5009–5010. [Google Scholar]
- 3.Cox H, Hargreaves S, Ismailov G. Effect of multi- drug resistance on global tuberculosis. The Lancet. 2003;362:1858–1859. doi: 10.1016/S0140-6736(03)14918-5. [DOI] [PubMed] [Google Scholar]
- 4.Winder FG, Collins PB. Inhibition by isoniazid of synthesis of mycolic acids in Mycobacterium tuberculosis. . J. Gen. Microbiol. 1970;63:41–48. doi: 10.1099/00221287-63-1-41. [DOI] [PubMed] [Google Scholar]
- 5.Winder FG, Collins PB, Rooney SA. Effects of isoniazid on mycolic acid synthesis in Mycobacterium tuberculosis and on its cell envelope. Biochemical J. 1979;117:127P. doi: 10.1042/bj1170027pa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole ST, Alzari PM. Towards new tuberculosis drugs. Biochem.Soc.Trans. 2007;35:1321–1324. doi: 10.1042/BST0351321. [DOI] [PubMed] [Google Scholar]
- 7.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 8.Marrakchi H, Lanéelle G, Quémard A. InhA, a target of the anti-tuberculosis drug isoniazid, is involved in a mycobacterial fatty acid elongation system. FAS-II. Microbiology. 2000;146:289–296. doi: 10.1099/00221287-146-2-289. [DOI] [PubMed] [Google Scholar]
- 9.Reche P, Perham RN. Structure and selectivity in post-translational modification : attaching the biotinyl – lysine and lipolyl- lysine swinging arms in multifunctional enzymes. EMBO J. 1999;18:2673–2682. doi: 10.1093/emboj/18.10.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reche P, Li YL, Fuller C, Eichhorn K, Perham RN. Selectivity of post-translational modification in binotinylated proteins: the carboxyl carrier protein of acetyl-CoA carboxylase of E.coli. Biochem. J. 1998;329:589–596. doi: 10.1042/bj3290589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman-Smith A, Cronan JE. The enzymatic biotinylation of proteins: a post- translational modification of exceptional specificity. Trends. Biochem. Soc. 1999;24:359–363. doi: 10.1016/s0968-0004(99)01438-3. [DOI] [PubMed] [Google Scholar]
- 12.Chapman- Smith A, Cronan JE. Molecular biology of biotin attachment to proteins. J. Nutr. 1999;129:477S–482S. doi: 10.1093/jn/129.2.477S. [DOI] [PubMed] [Google Scholar]
- 13.Mukhopadhyay B, Purwantini E, Kreder CL, Wolfe RS. Oxaloacetate synthesis in the Methanarchaeon Methanosarcina barkeri : Pyruvate carboxylase genes and a putative E.coli- type bifunctional biotin protein ligase gene (bpl/birA) exhibit a unique organization. J. Bac. 2001;183:3802–3810. doi: 10.1128/JB.183.12.3804-3810.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi -Rhee E, Cronan JE. The biotin carboxylase – biotin carboxylase carrier protein complex of E.coli acetyl-CoA carboxylase. J. Biol.Chem. 2003;278:30806–30812. doi: 10.1074/jbc.M302507200. [DOI] [PubMed] [Google Scholar]
- 15.Chapman-Smith A, Morris TW, Wallace JC, Cronan JE. Molecular recognition in a post-translational modification of exceptional specificity. Mutants of the biotinylate domain of acetyl- CoA caboxylase defective in recognition by biotin protein ligase. J. Biol. Chem. 1999;274:1449–1457. doi: 10.1074/jbc.274.3.1449. [DOI] [PubMed] [Google Scholar]
- 16.Xu Y, Beckett D. Evidence for interdomain interaction in the E.coli repressor of biotin biosynthesis from the studies of an N-terminal domain deletion mutant. Biochemistry. 1996;35:1783–1792. doi: 10.1021/bi952269e. [DOI] [PubMed] [Google Scholar]
- 17.Brown PH, Cronan JE, Grøti M, Beckett D. The biotin repressor: modulation of allostery by co-repressor analogs. J. Mol. Biol. 2004;337:857–869. doi: 10.1016/j.jmb.2004.01.041. [DOI] [PubMed] [Google Scholar]
- 18.Weaver LH, Kwon K, Beckett D, Mathews BW. Co-repressor induced organization and assembly of the biotin repressor: a model for allosteric activation of a transcriptional regulator. Proc. Natl. Acad. Sci. USA. 2001;98:6045–6050. doi: 10.1073/pnas.111128198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson JD, Higgins DG, Gibson TJ. Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions- specific gap penalties and weight matrix choice. Nucleic Acids. Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon K, Streaker ED, Beckett D. Binding specificity and the ligand dissociation process in the E.coli biotin holoenzyme synthetase. Protien Sci. 2002;11:558–570. doi: 10.1110/ps.33502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood ZA, Weaver LH, Brown PH, Beckett D, Mathews BW. Co- repressor induced order and biotin repressor dimerization : a case for divergent followed by convergent evolution. J. Mol. Biol. 2006;357:509–523. doi: 10.1016/j.jmb.2005.12.066. [DOI] [PubMed] [Google Scholar]
- 22.Bagautdinov B, Kuroishi C, Sugahara M, Kunishima N. crystal structure off biotin protein ligase from Pyrococcus horikoshii OT3 and its complexes: structural basis of biotin activation. J. Mol. Biol. 2005;353:322–333. doi: 10.1016/j.jmb.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 23.Kwon K, Beckett D. Function of a conserved sequence motif in biotin holoenzyme synthetase. Protein Sci. 2000;8:1530–1539. doi: 10.1110/ps.9.8.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown PH, Beckett D. Use of binding enthalpy to drive an allosteric transition. Biochemistry. 2005;44:3112–3121. doi: 10.1021/bi047792k. [DOI] [PubMed] [Google Scholar]
- 25.Xu Y, Beckett D. Thermodynamic analysis of small ligand binding to the E.coli repressor of biotin biosynthesis. Biochemistry. 1996;35:5509–5517. doi: 10.1021/bi9600658. [DOI] [PubMed] [Google Scholar]
- 26.Xu Y, Beckett D. Evidence for distinct ligand – bound conformational states of the multifunctional E.coli repressor of biotin biosynthesis. Biochemistry. 1995;343:16624–16631. doi: 10.1021/bi00051a010. [DOI] [PubMed] [Google Scholar]
- 27.Schrag JD, Bergeron JJ, Li Y, Borisova S, Hahn M, Thomas DY, Cygler M. The structure of calnexin, an ER chaperone involved in quality control of protein folding. Mol. Cell. 2001;8:633–644. doi: 10.1016/s1097-2765(01)00318-5. [DOI] [PubMed] [Google Scholar]
- 28.Polyak SW, Chapman- Smith A, Brautigan PJ, Wallace JS. Biotin protein ligase from Saccharomyces cerevisiae. The N-terminal domain is required for complete activity. J. Biol. Chem. 1999;274:32847–32854. doi: 10.1074/jbc.274.46.32847. [DOI] [PubMed] [Google Scholar]
- 29.Streaker ED, Beckett D. The biotin regulatory system: kinetic control of a transcriptional switch. Biochemistry. 2006;45:6417–6425. doi: 10.1021/bi052599r. [DOI] [PubMed] [Google Scholar]
- 30.Streaker ED, Beckett D. Coupling of protein assembly and DNA binding : biotin repressor dimerization precedes biotin operator binding. J. Mol. Biol. 2003;325:937–948. doi: 10.1016/s0022-2836(02)01308-6. [DOI] [PubMed] [Google Scholar]
- 31.Wilson KP, Shewchuk LM, Brennan RG, Ostuka AJ, Mathew BW. E.coli biotin holoenzyme syntheatase/ bio-repressor crystal structure delineates the biotin and DNA- binding domains. Proc. Natl. Acad. Sci. USA. 1992;89:9257–9261. doi: 10.1073/pnas.89.19.9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clarke DJ, Joseph C, Baillie R, Campopiano DJ. Biotinylation in the hyperthermophile Aquifes aeolicus. Isolation of a cross- linked BPL: BCCP complex. Eur. J. Biochem. 2003;270:1277–1287. doi: 10.1046/j.1432-1033.2003.03493.x. [DOI] [PubMed] [Google Scholar]
- 33.Wu S-C, Wong S-L. Development of an enzymatic method for site- specific incorporation of desthiobiotin to recombinant protein in vitro. . Anal. Biochem. 2004;331:340–348. doi: 10.1016/j.ab.2004.03.056. [DOI] [PubMed] [Google Scholar]
- 34.Lane DM, Rominger KL, Young DL, Lyen F. The enzymatic synthesis of holo- transcarboxylase from apo-carboxylase and (+) − biotin. J. Biol. Chem. 1964;239:2865–2871. [PubMed] [Google Scholar]
- 35.Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structures of biotin, bio-5′AMP and desthiobiotin. The ligand atoms are numbered as per the LPC/CSU software.
(0.05 MB DOC)