Abstract
The activity of the M26 meiotic recombination hot spot of Schizosaccharomyces pombe depends on the presence of the heptamer 5′-ATGACGT-3′. Transplacement of DNA fragments containing the ade6-M26 gene to other chromosomal loci has previously demonstrated that the heptamer functions in some, but not all, transplacements, suggesting that hot spot activity depends on chromosomal context. In this study, hot spot activity was tested in the absence of gross DNA changes by using site-directed mutagenesis to create the heptamer sequence at novel locations in the genome. When created by mutagenesis of 1–4 bp in the ade6 and ura4 genes, the heptamer was active as a recombination hot spot, in an orientation-independent manner, at all locations tested. Thus, the heptamer sequence can create an active hot spot in other chromosomal contexts, provided that the gross chromosomal structure is not altered; this result is consistent with the hypothesis that a specific higher-order chromatin structure is required for M26 hot spot activity.
Homologous recombination, the exchange of genetic information between homologous DNA duplexes, plays a vital role in the generation of genetic diversity, in the proper segregation of chromosomes during meiosis, and in the repair of DNA damage in somatic cells. Although homologous recombination occurs throughout the genome, some sites, termed hot spots, exhibit elevated frequencies of exchange. Recombination hot spots have been described in diverse organisms from bacteria to mammals (1, 2). Because hot spots appear to enhance a rate-limiting step of recombination, determination of the molecular mechanism by which hot spots act will increase our understanding of an important stage of homologous recombination.
The M26 mutation of the fission yeast Schizosaccharomyces pombe creates a meiotic recombination hot spot in the ade6 gene. M26 is one of 394 mutations in ade6 described by Gutz (3) and results from a G → T transversion in the coding region of the ade6 gene (4, 5). Among these 394 mutations, M26 is unique; in heteroallelic crosses, it recombines with other ade6 mutations up to 15-fold more frequently than the adjacent M375 mutation does (3). M375 is an equivalent G → T transversion in the preceding codon and thus serves as an ideal genetic control for M26 (4). M26 undergoes gene conversion 10 times more frequently than M375 and demonstrates disparity of conversion, with preferential conversion of M26 to wild type, whereas M375 converts with near parity (3). M26 also increases the frequency of conversion of other ade6 mutations, with which it frequently coconverts; the frequency of coconversion decreases with increasing distance from M26 (6). These properties led Gutz (3) to propose that the M26 mutation creates a site that stimulates recombination near itself and, less frequently, at a distance.
M26 is unique among eukaryotic hot spots in that hot spot activity is known to depend on a specific DNA sequence, the heptamer 5′-ATGACGT-3′, the first T in the heptamer sequence being the site of mutation in ade6-M26. Extensive mutational analysis has demonstrated that this heptamer is required for hot spot activity; mutation of any of the seven nucleotides results in reduction or abolition of hot spot activity, whereas mutations outside the heptamer sequence have little or no effect (7). A heterodimeric protein, Mts1/Mts2, binds specifically to the heptamer, and binding correlates with hot spot activity for a range of mutated substrates (8). Furthermore, M26 hot spot activity is abolished in mutants lacking this protein (W. Wahls, personal communication). Although M26 functions as a hot spot only in meiosis (5, 9), Mts1/Mts2 binding activity is present in extracts from mitotic as well as meiotic cells (8). Thus, although Mts1/Mts2 is necessary for recombination at M26, other factors appear to confer meiotic specificity.
The M26 heptamer is not the only chromosomal element necessary for hot spot activity. In recombination between the chromosomal ade6 locus and a plasmid carrying the ade6 gene, M26 is active when located on the chromosome but inactive when on the plasmid, implying that additional factors are required for hot spot activity (10, 11). The additional factor may be a second DNA sequence element or a specific chromatin structure in the region of ade6. Analysis of fragments of the ade6 gene bearing either M26 or the control M375 transplaced to various regions of the genome indicates that the heptamer can function as a hot spot in some, but not all, other locations (11). Hot spot activity was found to vary unpredictably when the size and orientation of the transplaced DNA was altered; for example, among insertions into ura4, a 4.4-kb fragment was active in two of four insertions, whereas a 3-kb fragment was active in one of six insertions. These observations suggest that the activity of the M26 hot spot is sensitive to its chromosomal context. In a separate study, deletion of the ade6 promoter abolished M26 hot spot activity (12), possibly reflecting the dependence of M26 on chromosomal context or, alternatively, a requirement for transcription.
We hypothesized that the M26 heptamer would be active as a recombination hot spot in other genomic locations, provided that the gross chromosomal structure is not changed. To test this hypothesis, we created the heptamer at novel genomic sites through site-directed mutagenesis of 1–4 bp of the wild-type sequence. This approach was taken to avoid gross alterations of chromosomal structure, in contrast to previous studies in which large regions of the ade6 gene were transplaced or deleted. We demonstrate that the heptamer is active as a hot spot in an orientation-independent manner at several sites in the ade6 gene and in either orientation in the ura4 gene. These results indicate that the M26 heptamer can function in a position- and orientation-independent manner, provided that the gross chromosomal structure is not changed, and, combined with other studies (11, 13), they support a role for chromatin structure in the regulation of hot spot activity in S. pombe.
MATERIALS AND METHODS
S. pombe Strains and Crosses.
The strains used for mutagenesis were GP18 (h− leu1–32) and GP20 (h+ leu1–32). Test alleles spanning the ade6 gene (see Fig. 1) have been described (5, 6). The ura4-595 allele is a duplication of 5′-GATC-3′ at nucleotide 595 (J. Bedoyan, personal communication). The ura4-294 allele is a C1212T transition (unpublished data). Strain genealogies are available upon request. All strains were maintained on YEA solid or YEL liquid media (14) supplemented with adenine (100 μg/ml) or on modified EMM2 minimal media (15) supplemented as appropriate. The frequency of heteroallelic recombination between ade6 or ura4 alleles was determined by random spore analysis of meiotic crosses (5). Spores were plated on EMM2 with or without adenine (or uracil) to assay total spore yield or Ade+ (or Ura+) recombinants, respectively. Each cross was repeated from independent cultures at least three times, and more than 50 colonies were counted for each determination. The frequency of Ade+ or Ura+ revertants, determined by meiotic selfings, was less than 0.1 per 106 viable spores (data not shown; 6, 16).
Figure 1.
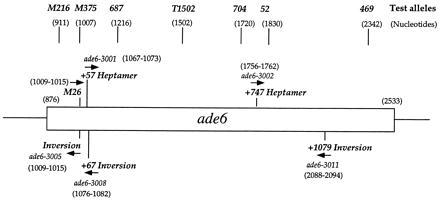
Map of the S. pombe ade6 gene showing positions of heptamer alleles generated in this study and test alleles. The orientation of the heptamer 5′-ATGACGT-3′ is indicated by arrows (5′ → 3′). The open rectangle indicates the coding region of ade6, nucleotides 876-2533 (4).
Chromosomal Mutagenesis.
Heptamer and control sequences were created in the chromosomal ade6 and ura4 loci by using site-directed mutagenesis and homologous recombination. All mutations were initially constructed in a plasmid and were then transferred to the chromosome by transforming strains GP18 or GP20 to either adenine or uracil auxotrophy with a linear fragment carrying the desired mutation. The mutations generated and their corresponding allele numbers are given in Table 1.
Table 1.
S. pombe ade6 and ura4 alleles
| Allele | Nucleotide(s) mutated* | Description |
|---|---|---|
| ade6-3001 | G1007T, A1068T, A1071C | Heptamer at 1067–1073 (+57 Heptamer) |
| ade6-3002 | T1759A, G1760C | Heptamer at 1756–1762 (+747 Heptamer) |
| ade6-3005 | G1007T, G1010C, A1012T, G1014A | Inverted heptamer at 1009–1015 (Inversion) |
| ade6-3006 | G1007T, A1012T, G1014A, G1016C | Control for ade6-3005 |
| ade6-3007 | T1759A, G1761C | Control for ade6-3002 |
| ade6-3008 | T1080C, T1083A | Inverted heptamer at 1076–1082 (+67 Inversion) |
| ade6-3009 | T2097A | Control for ade6-3011 |
| ade6-3010 | T1079C, T1083A | Control for ade6-3008 |
| ade6-3011 | C2088A | Inverted heptamer at 2088–2094 (+1079 Inversion) |
| ura4-167 | G959A, G960T | Heptamer at 959–965 |
| ura4-168 | G960T, G967A | Control for ura4-167 |
| ura4-169 | G966C, G967A, T973A | Inverted heptamer at 962–968 |
| ura4-170 | G960C, G961A, T973A | Control for ura4-169 |
The ade6-3001 and ade6-3002 alleles were generated by creation of mutations in a plasmid template using Esherichia coli strain CJ236 [dut-1 ung-1 relA1 (pCJ105)] as described by Kunkel (18). GP20 was transformed to adenine auxotrophy with a 1.45-kb BamHI/XhoI fragment to give ade6-3001. GP18 was transformed to adenine auxotrophy with a 1.96-kb BamHI/SpeI fragment to give ade6-3002.
All other alleles were generated by PCR-based mutagenesis as follows. Complementary PCR primers containing the desired mutations were used in combination with external primers to amplify two products containing an overlapping region of mutated DNA. These products were combined, melted, and re-amplified by using the external primer pair. All PCRs contained 10 ng of template plasmid DNA, 50 pmol of each primer, 1.5 mM MgCl2, 250 μM dNTPs, and 0.5 units of Taq polymerase (Promega) in the supplied reaction buffer (50-μl final volume) and were performed under the following conditions: 94° for 30 s, 55° for 30 s, 72° for 30 s, with 30 cycles of amplification. The product of the secondary amplification was subcloned into pCRII (Invitrogen) and sequenced to confirm that only the desired mutations were present. A restriction fragment containing the mutated sequence was used to replace the same region in a plasmid containing the full-length wild-type ade6 or ura4 gene. A 1.45-kb BamHI/XhoI fragment of ade6 or a 1.75-kb HindIII fragment of ura4 was used to transform strain GP18 to adenine or uracil auxotrophy, respectively.
S. pombe was transformed with 1 μg of a linear DNA fragment carrying the desired mutation by using lithium acetate as described (19). Fragments containing ade6 mutations were cotransformed with plasmid pDB248′ (20) carrying the Saccharomyces cerevisiae LEU2 gene, and Leu+ transformants were selected on EMM2 plates containing no leucine and limiting adenine (10 μg/ml) to allow identification of adenine auxotrophs by their red color. Because integration of the cotransforming plasmid is possible, all strains were purified on medium containing leucine, and Leu− segregants were identified. Uracil auxotrophs were selected on YEA medium containing 5-fluoroorotic acid (1 mg/ml). Each mutation was confirmed by PCR amplification and sequencing of genomic DNA, and by Southern blot analysis of genomic DNA digests (data not shown).
All mutations were designed to include the base changes required to generate the M26 heptamer and an additional mutation that generates a stop codon to ensure that the strains are auxotrophic. For ade6-3001, the nonsense codon was provided by the M375 mutation (Table 1). For all other strains in which a heptamer was generated, a control strain in which the same number and type of base changes were made without generating the heptamer was designed (Table 1). The strain carrying allele ade6-3011 was found to contain a single mutation that creates the heptamer without the accompanying nonsense mutation. This strain was auxotrophic for adenine and produced dark red colonies on media containing limiting adenine. The structure of the ade6 gene appeared normal by Southern blot analysis, and no additional mutations were found after sequencing 1.5 kb of the coding region. The control strain (allele ade6-3009) carries a single nonsense mutation 9 bp from the ade6-3011 mutation.
RESULTS
The Heptamer Functions as a Hot Spot When Moved Within the ade6 Gene.
To determine whether the heptamer was active as a hot spot when moved within the ade6 gene, we used site-directed mutagenesis to create the heptamer at two locations downstream (with respect to the direction of transcription) of the site of M26. The new locations were selected to minimize the number of bases mutated to generate the heptamer. The mutation ade6-3001 creates the heptamer 57 bp downstream of M26 (designated the +57 Heptamer), and ade6-3002 creates the heptamer 747 bp downstream of M26 (+747 Heptamer; Fig. 1). Both strains are wild-type at the M26 site. Because the +57 Heptamer was constructed in a background bearing the M375 mutation, the control used was M375. Recombinant frequencies were determined by crossing strains bearing heptamers or the corresponding control alleles with six test strains carrying alleles that span the coding region of the ade6 gene (Fig. 1). Except for the +57 Heptamer and +1079 Inversion (see Materials and Methods) the control alleles contain the same number and type of base pair changes as the corresponding heptamer does. The recombinant frequency increased with increasing distance between the alleles (Fig. 2), as shown previously for M26 (3, 6). The presence of the heptamer at both the +57 and +747 sites increased recombination relative to the control when crossed with each of the test strains. The hot spot activity, defined as the ratio of the recombinant frequency with the heptamer to that with the control mutation, was 4.0–7.5 for the +57 Heptamer and 6.7–14.0 for the +747 Heptamer, depending upon the test allele (Table 2). The hot spot activity of M26, relative to M375, when crossed with the same series of markers was 5–15 (Fig. 2A and Table 2), in close agreement with other studies (3, 6). Hot spot activity of the +747 Heptamer was therefore comparable to that of M26, whereas activity of the +57 Heptamer was somewhat lower. Thus, although the heptamer was active at both new sites tested within the ade6 gene, the level of activity may depend upon the position of the heptamer within the gene (see Discussion).
Figure 2.
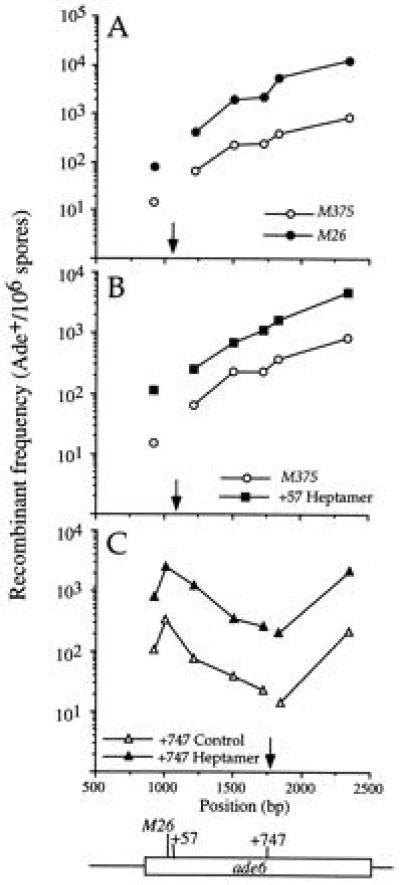
The heptamer is active when moved to new locations within the ade6 gene. (A) The original M26 heptamer. (B) The heptamer moved 57 bp downstream of M26 (+57 Heptamer; ade6-3001). (C) The heptamer moved 747 bp downstream of M26 (+747 Heptamer; ade6-3002). Each heptamer allele was crossed with a series of test alleles, and the recombinant frequency (determined as the number of Ade+ spores per 106 viable spores) is plotted against the position of the test allele along the ade6 gene. The open rectangle indicates the coding region of ade6 (Fig. 1). Closed symbols represent heptamer mutations, and open symbols indicate control strains that contain the same number and type of base changes without creating the heptamer. The arrow indicates the position of the heptamer sequence.
Table 2.
Hot spot activity of heptamers created within the ade6 gene
| ade6 test allele | Heptamer allele
|
|||||
|---|---|---|---|---|---|---|
| ade6-M26 | ade6-3001 | ade6-3002 | ade6-3005 | ade6-3008 | ade6-3011 | |
| M26 | +57 Heptamer | +747 Heptamer | Inversion | +67 Inversion | +1079 Inversion | |
| M216 | 5.1 | 7.5 | 6.7 | 19.7 | 4.0 | 18.4 |
| [2.8–7.0 (5)] | [5.3–9.7 (4)] | [3.9–9.0 (3)] | [11.5–29.3 (6)] | 3.3–5.0 (4)] | [10.2–28.7 (6)] | |
| M375 | ND | ND | 7.7 | ND | 3.7 | 20.1 |
| [3.8–12.3 (7)] | [2.5–6.3 (4)] | [12.4–28.1 (6)] | ||||
| 687 | 7.0 | 4.0 | 8.6 | 10.6 | 6.1 | 19.3 |
| [5.1–9.3 (3)] | [2.7–5.2 (3)] | [4.2–12.3 (4)] | 6.8–13.2 (4)] | [4.3–10.4 (4)] | [15.9–23.7 (4)] | |
| T1502 | 9.3 | 4.9 | 8.0 | 13.4 | 5.9 | 9.3 |
| [8.5–10.5 (3)] | [2.5–10.0 (4)] | [4.8–10.5 (4)] | [7.4–19.6 (4)] | [3.1–8.0 (3)] | [7.7–11.6 (3)] | |
| 704 | 9.2 | 4.8 | 14.0 | 12.5 | 7.1 | 12.5 |
| [6.5–11.1 (3)] | [3.5–8.1 (4)] | [10.4–18.1 (4)] | [8.5–16.3 (4)] | [4.1–10.7 (3)] | [9.8–17.8 (3)] | |
| 52 | 14.0 | 4.3 | 11.9 | 25.3 | 11.6 | 2.9 |
| [9.0–24.0(7)] | [3.8–4.9 (4)] | [5.2–16.2 (3)] | [18.8–26.6 (7)] | [7.6–17.2 (5)] | [1.4–6.1 (5)] | |
| 469 | 15.0 | 4.0 | 8.6 | 19.1 | 9.6 | 7.3 |
| [14.3–21.5 (3)] | [2.6–5.1 (4)] | [5.6–12.8 (4)] | [13.7–25.5 (4)] | [6.2–14.9 (5)] | [2.4–12.5 (6)] | |
Hot spot activity is the recombinant frequency for the heptamer allele divided by the recombinant frequency for the corresponding control allele, each crossed with the indicated test allele. Hot spot activity is given as the mean value. The range is shown in brackets along with the number of independent crosses analyzed (n). ND, not determined.
The Heptamer Functions as a Hot Spot in the Inverted Orientation Within the ade6 Gene.
Because hot spot activity of M26 depended on the orientation of certain transplacements of the ade6 gene into the ura4 locus (11), we tested the orientation-dependence of heptamers made by site-directed mutagenesis within ade6. Strains were constructed with the heptamer sequence in an inverted orientation relative to M26 at three positions within the ade6 gene; at the original M26 site and 67 bp or 1079 bp downstream of the original site (alleles ade6-3005, -3008, and -3011, designated Inversion, +67 Inversion, and +1079 Inversion, respectively; Fig. 1 and Table 1). Strains carrying the inversions, or the corresponding controls, were crossed with the test strains, and the recombinant frequencies and hot spot activities were determined as above (Fig. 3 and Table 2). The inverted heptamer was active at all three locations, with the hot spot activity varying from 2.9 to 25.3. Hot spot activity was generally higher for the Inversion at the M26 site (12.5–25.3) than for the +1079 Inversion (2.9–20.1) or the +67 Inversion (3.7–11.6).
Figure 3.
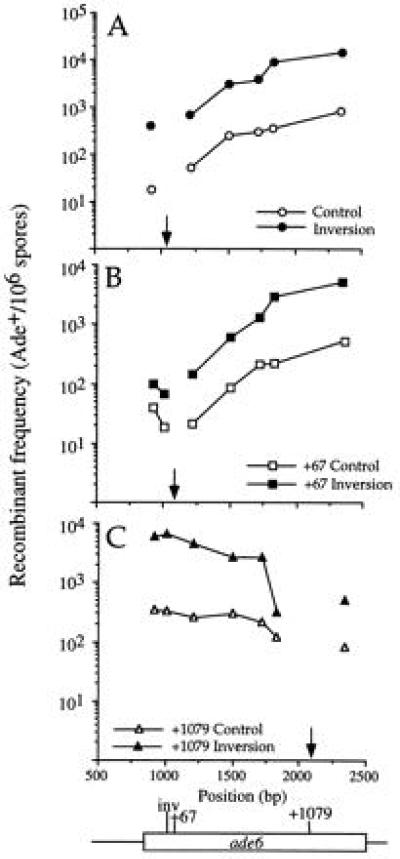
The heptamer is active in the inverted orientation within the ade6 gene. (A) Inversion at the site of M26 (Inversion; ade6-3005). (B) Inverted heptamer 67 bp downstream of M26 (+67 Inversion; ade6-3008). (C) Inverted heptamer 1079 bp downstream of M26 (+1079 Inversion; ade6-3011). See Fig. 2 legend for explanation of symbols.
Because heteroduplex DNA is formed as an intermediate in homologous recombination (reviewed in refs. 21 and 22), the efficiency of repair of mismatched bases may influence levels of recombination. However, the recombinant frequencies for the inversion control, which contains four mutated bases, were comparable to those of the adjacent M375 allele, which contains a single base pair change (4), indicating that the number of mismatches does not significantly influence levels of recombination in these crosses.
The Heptamer Functions as a Hot Spot in Both Orientations Within the ura4 Gene.
The heptamer sequence was active at all locations tested within the ade6 gene in a position- and orientation-independent manner. To investigate the possibility that hot spot activity was restricted to the ade6 gene, we constructed strains with the heptamer sequence in either orientation at one site within the coding region of the ura4 gene. Recombinant frequencies were determined from heteroallelic crosses with strains bearing the test alleles ura4-595 or ura4-294, which map to either side of the heptamer mutations. The heptamer was active as a hot spot in each orientation, although activity was slightly higher in one orientation (7.5–8.1) than in the other (4.8–5.1; Table 3). The ability of the heptamer to function as a recombination hot spot is therefore not restricted to recombination within the ade6 gene.
Table 3.
Recombinant frequency and hot spot activity for heptamers created within the ura4 gene
| Heptamer allele | Test allele | Recombinant frequency (Ura+ per 106 spores) | Hot spot activity* |
|---|---|---|---|
| Heptamer in ura4 | |||
| ura4-167 (heptamer) | ura4-294 | 965, 768, 488, 679, 700 | 8.1 [3.4–17.5 (5)] |
| ura4-168 (control) | ura4-294 | 55, 87, 85, 202, 141 | NA |
| ura4-167 (heptamer) | ura4-595 | 1017, 703, 615, 1469, 927, 633 | 9.5 [4.7–19.8 (6)] |
| ura4-168 (control) | ura4-595 | 182, 121, 31, 160, 196, 54 | NA |
| Inversion in ura4 | |||
| ura4-169 (inversion) | ura4-294 | 138, 156, 190, 140, 136 | 4.8 [2.6–8.0 (5)] |
| ura4-170 (control) | ura4-294 | 54, 31, 24, 46, 25 | NA |
| ura4-169 (inversion) | ura4-595 | 227, 205, 237, 142, 159 | 5.3 [2.9–10.7 (5)] |
| ura4-170 (control) | ura4-595 | 78, 60, 22, 28, 35 | NA |
Hot spot activity is the recombinant frequency for the cross with the heptamer divided by that for the corresponding control. Crosses were performed in pairs, in the order indicated. Mean values are given; the range and number of independent crosses (n) performed are given in brackets. NA, not applicable.
DISCUSSION
Recombination hot spot activity of the ade6-M26 mutation requires the heptamer sequence 5′-ATGACGT-3′ (7). We demonstrate here that this heptamer functions as a hot spot in an orientation-independent manner when moved to new locations in the ade6 gene and in the ura4 gene. Thus, other sites in ade6 and other loci in the S. pombe genome are receptive to an active recombination hot spot provided by the M26 heptamer sequence.
The M26 heptamer is not, however, the only chromosomal element required for hot spot activity. It has been proposed that either an additional DNA sequence or a particular chromatin context is necessary (10, 11). We have now demonstrated activity of the heptamer in several sites in the genome, indicating that a requirement for an additional specific DNA sequence is unlikely. However, our results are consistent with the hypothesis that a specific chromatin context is required for hot spot activity. Because M26-dependent hot spot activity varies in an unpredictable fashion with transplacements of different ade6 fragments to ura4, and in some cases shows position- and orientation-dependence, we previously inferred that chromatin structure is important for M26 activity (11). These results are similar to those of studies in Saccharomyces cerevisiae, in which recombination hot spot activity varies in a nonsystematic manner after transplacemant of a hot spot region from one chromosomal locus to another (reviewed in ref. 2). All of these studies involved transplacements of relatively large fragments of DNA (usually >1 kb), often accompanied by plasmid DNA at the junction with the chromosomal integration site. We hypothesize that integrations of this size, combined with the creation of new sequences at the junctions, alter the gross chromosomal structure and account for the unpredictable M26 hot spot activities. In this study, we have made base changes of one to four nucleotides to move the heptamer to new locations within the ade6 and ura4 genes, without changing the spacing between chromosomal elements and with the aim of minimizing disruptions to chromatin structure. We find that the heptamer is active as a recombination hot spot, in an orientation-independent manner, at all five locations tested. These studies demonstrate that the heptamer can create a hot spot in different chromosomal contexts and is inactive only when certain large chromosomal integrations are made.
Several lines of evidence support a role for chromatin structure in hot spot activity. In Saccharomyces cerevisiae, there is convincing evidence that meiotic recombination proceeds through the formation of transient double-strand breaks (DSBs) (23–27). Sites at which DSBs are formed are hypersensitive to nuclease digestion of isolated chromatin, and changes in chromatin structure that alter nuclease sensitivity also result in altered DSB patterns (28, 29). However, other factors, such as competition between closely spaced DSB sites, may influence hot spot activity because nuclease hypersensitivity is not sufficient for DSB formation in all contexts (30). It appears that in Saccharomyces cerevisiae local chromatin structure in early meiosis plays an important role in determining the position of DSBs and hence the initiation of recombination. The demonstration of nuclease-sensitive sites at a meiotic recombination hot spot in the Eβ gene of the major histocompatibility locus in mouse (31) indicates that chromatin structure may be a general factor influencing meiotic recombination in eukaryotes.
We propose that the M26 heptamer and associated binding proteins, including Mts1/Mts2 (8), interact with a specific chromatin structure to promote recombination. The binding of Mts1/Mts2 to the heptamer may induce an appropriate chromatin structure, or binding may follow formation of a favorable chromatin structure at the heptamer sequence. Induction of this structure could be inhibited in some chromosomal contexts when gross structural changes are involved, possibly by inducing antagonistic chromatin structures or by altering the spacing between determinants of nucleosome positioning.
There is physical evidence that chromatin structure plays a role in M26 recombination hot spot activity. A micrococcal nuclease-sensitive site has been detected near M26; this site is present in chromatin from ade6-M26 cells but is absent in both ade6-M375 and ade6+ chromatin (13). Furthermore, in strains bearing single base pair changes in the vicinity of M26 (7), the presence of the nuclease-sensitive site in isolated chromatin correlates both with binding of the purified Mts1/Mts2 protein to DNA and with meiotic hot spot activity (8, 13). This correlation of hot spot activity, Mts1/Mts2 binding, and nuclease sensitivity supports the hypothesis that chromatin structure is important for M26 activity.
Although the heptamer was active at all locations tested within the ade6 gene, the level of hot spot activity varied from 3 to 25 (Table 2). The highest levels of activity were attained for the Inversion at the M26 site, the +1079 Inversion, and the +747 Heptamer; in the Inversion and +1079 Inversion, the heptamer sequence is followed by a C residue, and in the +747 Heptamer, the heptamer is preceded by a T residue. The sequences 5′-ATGACGTC-3′ and 5′-TATGACGT-3′ are present at the site of M26 in the G1016C and G1008T mutants described by Schuchert et al. (7). These mutations each increased the hot spot activity of M26 about 1.5-fold. The variation in hot spot activity between heptamers created at different sites within ade6 may therefore be explained by differences in the residues adjacent to the heptamer.
This study demonstrates that a eukaryotic recombination hot spot can be moved with a high degree of flexibility with regard to its position and orientation, similar to the findings with many transcriptional enhancers. A critical nucleotide sequence and similar flexibility of movement have not been demonstrated for other eukaryotic recombination hot spots. The heptamer sequence is necessary for M26 activity (7). We now show that, with regard to primary DNA sequence, the heptamer sequence does not require a fixed relation to any other DNA element. The ability to move the heptamer and retain activity provides the potential to create a recombination hot spot at any site in the genome and will be a powerful tool in future studies of the interaction between recombination hot spots and chromatin structure.
Acknowledgments
We thank Yukang Lin for PCR mutagenesis to create alleles ade6-3001 and ade6-3002; Jirair Bedoyan for sequencing the ura4-595 mutation; the Fred Hutchinson Biotechnology Center for DNA sequencing and oligonucleotide synthesis; Kunihiro Ohta and Wayne Wahls for sharing unpublished data; David Beach and Jürg Kohli for S. pombe strains; and Sue Amundsen, Jirair Bedoyan, Marcella Cervantes, Ramsay McFarlane, and Andrew Taylor for helpful discussion and comments on the manuscript. This research was supported by National Institute of General Medical Sciences Grant GM31693 to G.R.S. and by fellowships from the National Cancer Institute (CA08922) and the Cancer Research Foundation of America to J.B.V.
ABBREVIATION
- DSB
double-strand break
References
- 1.Smith G R. Experientia. 1994;50:234–241. doi: 10.1007/BF01924006. [DOI] [PubMed] [Google Scholar]
- 2.Lichten M, Goldman A S H. Annu Rev Genet. 1995;29:423–444. doi: 10.1146/annurev.ge.29.120195.002231. [DOI] [PubMed] [Google Scholar]
- 3.Gutz H. Genetics. 1971;69:317–337. doi: 10.1093/genetics/69.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szankasi P, Heyer W D, Schuchert P, Kohli J. J Mol Biol. 1988;204:917–925. doi: 10.1016/0022-2836(88)90051-4. [DOI] [PubMed] [Google Scholar]
- 5.Ponticelli A S, Sena E P, Smith G R. Genetics. 1988;119:491–497. doi: 10.1093/genetics/119.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zahn-Zabal M, Kohli J. Curr Genet. 1996;29:530–536. doi: 10.1007/BF02426957. [DOI] [PubMed] [Google Scholar]
- 7.Schuchert P, Langsford M, Käslin E, Kohli J. EMBO J. 1991;10:2157–2163. doi: 10.1002/j.1460-2075.1991.tb07750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wahls W P, Smith G R. Genes Dev. 1994;8:1693–1702. doi: 10.1101/gad.8.14.1693. [DOI] [PubMed] [Google Scholar]
- 9.Schuchert P, Kohli J. Genetics. 1988;119:507–515. doi: 10.1093/genetics/119.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ponticelli A S, Smith G R. Proc Natl Acad Sci USA. 1992;89:227–231. doi: 10.1073/pnas.89.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Virgin J B, Metzger J, Smith G R. Genetics. 1995;141:33–48. doi: 10.1093/genetics/141.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zahn-Zabal M, Lehmann E, Kohli J. Genetics. 1995;140:469–478. doi: 10.1093/genetics/140.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizuno K-i, Emura Y, Baur M, Kohli J, Ohta K, Shibata T. Genes Dev. 1997;11:876–886. doi: 10.1101/gad.11.7.876. [DOI] [PubMed] [Google Scholar]
- 14.Gutz H, Heslot H, Leupold U, Loprieno N. In: Handbook of Genetics. King R C, editor. Vol. 1. New York: Plenum; 1974. pp. 395–446. [Google Scholar]
- 15.Nurse P. Nature (London) 1975;256:547–551. doi: 10.1038/256547a0. [DOI] [PubMed] [Google Scholar]
- 16.DeVeaux L C, Hoagland N A, Smith G R. Genetics. 1992;130:251–262. doi: 10.1093/genetics/130.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimm C, Kohli J, Murray J, Maundrell K. Mol Gen Genet. 1988;215:81–86. doi: 10.1007/BF00331307. [DOI] [PubMed] [Google Scholar]
- 18.Kunkel T A. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito H, Fukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beach D, Piper M, Nurse P. Mol Gen Genet. 1982;187:326–329. doi: 10.1007/BF00331138. [DOI] [PubMed] [Google Scholar]
- 21.Orr-Weaver T L, Szostak J W. Microbiol Rev. 1985;49:33–58. doi: 10.1128/mr.49.1.33-58.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith G R. Annu Rev Genet. 1987;21:179–201. doi: 10.1146/annurev.ge.21.120187.001143. [DOI] [PubMed] [Google Scholar]
- 23.Sun H, Treco D, Schultes N P, Szostak J W. Nature (London) 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- 24.Cao L, Alani E, Kleckner N. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- 25.Sun H, Treco D, Szostak J W. Cell. 1991;64:1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- 26.Padmore R, Cao L, Kleckner N. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- 27.Goyon C, Lichten M. Mol Cell Biol. 1993;13:373–382. doi: 10.1128/mcb.13.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohta K, Shibata T, Nicolas A. EMBO J. 1994;13:5754–5763. doi: 10.1002/j.1460-2075.1994.tb06913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu T-C, Lichten M. Science. 1994;263:515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- 30.Wu T-C, Lichten M. Genetics. 1995;140:55–66. doi: 10.1093/genetics/140.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shenkar R, Shen M H, Arnheim N. Mol Cell Biol. 1991;11:1813–1819. doi: 10.1128/mcb.11.4.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]


