Abstract
A mainstay of the antiretroviral drugs used for therapy of HIV-1, Zidovudine (AZT) is genotoxic and becomes incorporated into DNA. Here we explored host interindividual variability in AZT-DNA incorporation, by AZT radiomimmunoassay (RIA), using 19 different strains of normal human mammary epithelial cells (NHMECs) exposed for 24 h to 200 μM AZT. Twelve of the 19 NHMEC strains showed detectable AZT-DNA incorporation levels (16 to 259 molecules of AZT/106 nucleotides), while 7 NHMEC strains did not show detectable AZT-DNA incorporation. In order to explore the basis for this variability, we compared the 2 NHMEC strains that showed the highest levels of AZT-DNA incorporation (H1 and H2) with 2 strains showing no detectable AZT-DNA incorporation (L1 and L2). All 4 strains had similar (≥80%) cell survival, low levels of accumulation of cells in S-phase, and no relevant differences in response to the direct-acting mutagen bleomycin (BLM). Finally, when levels of thymidine kinase 1 (TK1), the first enzyme in the pathway for incorporation of AZT into DNA, were determined by Western blot analysis in all 19 NHMEC strains at 24 h of AZT exposure, higher TK-1 protein levels were found in the 12 strains showing AZT-DNA incorporation, compared to the 7 showing no incorporation (p = 0.0005, Mann-Whitney test). Furthermore, strains L1 and L2, which did not show AZT-DNA incorporation at 24 h, did have measurable incorporation by 48 and 72 h. These data suggest that variability in AZT-DNA incorporation may be modulated by interindividual differences in the rate of induction of TK1 in response to AZT exposure.
INTRODUCTION
The nucleoside reverse transcriptase inhibitor (NRTI), 3′-azido-3′-deoxythymidine (AZT) comprises part of the first-line therapy for HIV-1 infection worldwide (DHHS, 2006), and is specifically recommended by the Centers for Disease Control for inhibition of mother-to-child HIV-1 transmission (Centers for Disease Control and Prevention, 2003) and post-exposure prophylaxis in health care, laboratory and rescue workers (Cardo et al., 1997; Centers for Disease Control and Prevention, 1999). The genotoxicity of this drug has been studied extensively, and numerous reports indicate that incorporation of the drug into DNA, clastogenicity and mutagenicity are consequences of AZT exposure in cultured cells, animal models and humans (IARC, 2000; Poirier et al., 2004; Escobar et al., 2007; Olivero, 2007) Transplacental carcinogenesis in mouse offspring exposed to AZT during the last week of gestation was documented in multiple organs at 1–2 years of age (Olivero et al., 1997; Diwan et al., 1999; NTP, 1996; Walker et al., 2007). The studies revealed dose-related increases in incidences of liver, lung and reproductive organ tumors, and raised some concerns regarding potential cancer risk in human infants exposed to the NRTI drugs during development.
Mechanisms underlying human interindividual response to AZT may be important for considerations of drug efficacy and toxicity. Here we explore the genotoxicity of AZT in 19 mammary epithelial cell strains derived from human breast tissue taken from 19 donors at reduction mammoplasty. These normal human mammary epithelial cells (NHMECs) have been characterized previously (Keshava et al., 2005), and constitute an ideal model to assess human interindividual variability.
The antiretroviral nucleoside analog drug AZT becomes incorporated into nascent HIV-1 viral DNA via reverse transcriptase, and into the host DNA via classical polymerases. Prior to this incorporation the drug must be phosphorylated, by thymidine kinase 1 (TK1) followed by thymidylate kinase and nucleoside diphosphate kinase (Furman et al., 1986). Loss of TK1 activity has been observed in cultured cells exposed long-term to AZT (Avramis et al., 1993; Wu et al., 1995; Vazquez et al., 2004) and in patients exposed chronically to therapeutic AZT doses (Avramis et al., 1993). To understand interindividual variability in genotoxic insult and metabolic capacity, with the intention of elucidating clinical efficacy and resistance, we investigated AZT-DNA incorporation in a panel of 19 different NHMEC strains and correlated AZT-DNA incorporation values with protein levels of the enzymatically-active (24KD) TK1.
MATERIAL AND METHODS
Cell culture, AZT exposure and DNA preparation
Normal human mammary epithelial cells (NHMECs) were cultured from organoids derived from tissues obtained at reduction mammoplasty from 19 different individuals by the Cooperative Human Tissue Network. Cells were grown at 37°C in 5% CO2 and serum free Mammary Epithelial Cell Medium (Cambrex, Rockland, ME) supplemented with growth factors, insulin and pituitary extracts (Cambrex) (Keshava et al., 2005). The cells were grown out to passage 6 to remove stromal components. AZT (Sigma-Aldrich Co, St Louis, MO) was dissolved in phosphate buffered saline (PBS) pH 7.2 (Biosource, Rockville, MD) and the final AZT concentration was calculated from absorbance at 266 nm. NHMECs from 19 different individuals were cultured to passage 6 and exposed to 200 μM AZT for 24 h, rinsed twice with PBS, removed from the flask by trypsin (Cambrex) and treated with trypsin-neutralizing agent (Cambrex) before centrifugation. Cell pellets were washed with PBS and then processed for DNA extraction using phenol chloroform (Sambrook et al., 1989). This experiment was performed twice.
Measurement of AZT-DNA incorporation by AZT radioimmunoassay (RIA)
DNA quantity was determined by absorbance at 260 nm in DNA aliquots from 19 NHMEC strains exposed on two separate occasions to either to 0 or 200 μM AZT for 24 h. Samples were diluted to 30 μg DNA/ml using 10 mM Tris, 1 mM EDTA (TE) buffer, sonicated for 30 sec, warmed at 42°C for 15 min, heated at 99°C in a thermomixer (Eppendorf, Hamburg, Germany) for 2 min, and placed immediately on ice.
The incorporation of AZT into NHMEC-DNA was determined by AZT RIA (Olivero et al., 1994). Briefly, a rabbit polyclonal anti-AZT antibody (Sigma-Aldrich Co), which also recognizes AZT in DNA (Olivero et al., 1994), was reconstituted, diluted 1:7500, and incubated with NHMEC-DNA for 90 min at 37°C. An aliquot (100 μl) containing approximately 20,000 cpm of [3H]AZT tracer (16 Ci/mmol, Moravek Biochemicals Inc. Mountain View, CA) was added to each tube together with 100 μl of the secondary antibody, goat anti-rabbit immunoglobulin G (Sigma-Aldrich Co). The mixture was incubated for 25 min at 4°C, centrifuged at 3,000 rpm for 15 min at 4°C and the resulting supernatant was decanted. The pellets were dissolved in 0.1M NaOH and counted in a liquid scintillation counter. The amount of standard AZT, added to 3 μg of NHMEC control DNA, required to inhibit antibody binding by 50% was 176.7 ± 34.4 (average ± SD, n=5) molecules of AZT/106 nucleotides. The lower limit of detection was 16 molecules of AZT/106 nucleotides.
NHMEC DNA samples from the 19 strains, obtained from cells exposed to 200 μM AZT for 24 h on two separate occasions, were assayed in duplicate in 3 separate RIAs. In addition to the 24 h exposure, NHMEC L1 and L2 cells were exposed to 200 μM AZT for periods of 48 and 72 h, and incorporation of AZT into DNA was determined by RIA.
Cell viability, micronuclei and apoptosis in 4 NHMEC strains exposed to bleomycin
Selected end points were examined in a subset of 4 NHMEC strains. NHMEC strains M99005 and M98018, designated H1 and H2 respectively, incorporated the highest levels of AZT into DNA when measured by AZT-RIA, after 24 h of exposure to 200 μM AZT. NHMEC strains M98016 and M98040, designated L1 and L2 respectively, were two of the strains that incorporated undetectable levels of AZT into DNA when measured by AZT-RIA, after 24 h of exposure to 200 μM AZT. For cell survival studies, these 4 NHMEC strains were grown to semi-confluency (~ 80% cell growth) in 10 mm Petri dishes, at which time 3 dishes of cells of each strain were exposed to either 0 or 200 μM AZT for 24 h. The experiment was repeated twice. Dishes were rinsed twice with PBS, trypsinized, neutralized, and an aliquot was taken and counted twice in a Coulter Counter (Coulter Electronics Limited, England). Cell counts in drug-exposed cultures were compared with those in the unexposed dishes, and survival in exposed cells was expressed as percentage of survival in unexposed cells.
The four NHMEC strains were exposed for 4 h, on two separate occasions, to 36 μM Bleomycin (BLM, Sigma-Aldrich Co). Cells were rinsed with PBS, cultured in fresh media for an additional 20 h, trypsinized, collected by centrifugation and resuspended in media containing serum to produce spreads on slides. Cells were fixed with Carnoy’s fixative (3:1 methanol: glacial acetic acid), and stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) and evaluated microscopically. The terminal deoxynucleotidyl transferase (Tdt) mediated dUTP nick end labeling (TUNEL) assay was performed to evaluate apoptosis in strains H1, H2, L1 and L2 using the Apoptag, Fluorescein In Situ Apoptosis Detection kit (Intergen, Purchase, NY). Apoptotic figures were scored in cells exposed to 0 or 36 μM BLM for 4 h, and then incubated for 20 h with fresh media. Apoptotic figures were visualized and photographed using a Nikon Eclipse E-400 (Nikon, Inc, Melville, NY) microscope with fluorescence capabilities. Micronuclei were observed with the use of a UV-2E/C DAPI filter and excitation and barrier filters of 330–380 nm, and 400 nm respectively. Scoring of apoptotic figures was performed with the use of a triple pass filter with excitation filters of 390–402 and 478–495 for DAPI and FITC, respectively, the barrier filters used were of 462 and 523 nm for DAPI and FITC respectively.
Cell cycle analysis by flow cytometry
In two independent experiments NHMEC strains H1, H2, L1 and L2 were plated in six-well plates, and, for each plate, three wells were unexposed and three wells were exposed to 200μM AZT for 24 h. Cells (106) were harvested, pelleted and washed with culture medium without serum before they were fixed in ice-cold ethanol (1 ml; 70%) and dropped onto slides while vortexing. Following an overnight fixation at 4°C, cells were pelleted by centrifugation and incubated with Ribonuclease A (Sigma-Aldrich Co) at room temperature for 20 min. Propidium iodide (20–50 μg/ml) (Molecular Probes, Eugene, OR) was added to each cell suspension and cells were kept in the dark at 4°C overnight. Cells were passed through a fluorescence activated cell sorter (FACSCalibur, BD Biosciences, San Jose, CA) using the doublet discrimination module, and data were acquired using CellQuest (BD Biosciences) software. The cell cycle was modeled using ModFit software (Venty Software, Topsham, ME). Percentages of cells in G0-G1, S and G2-M phases were calculated directly by the software.
Thymidine kinase 1 (TK1) protein quantitation by Western Blot
Aliquots of cells from each of the 19 NHMEC strains were lysed in Radio-immune precipitation assay (RIPA) buffer (50 mM Tris-HCL pH 7.6, 150 mM NaCl, 0.25% SDS, 1% Triton X-100, 1 mM EDTA, 0.5% Nonidet P40) for 30 min on ice, followed by sonication for 30 sec. Proteins were quantified by Bradford reaction (Bio Rad, Hercules, CA). Samples were resolved on a 10% Bis-Tris vertical polyacrylamide gel (NuPage, Invitrogen, Carlsbad, CA) and then transferred to a nitrocellulose membrane. A TK1 monoclonal antibody (QED Biosciences, San Diego, CA) was used to incubate the membrane overnight at 4°C. After incubation with an anti-mouse IgG-horse radish peroxidase (HRP)-conjugated secondary antibody (Novus Biologicals, Littleton, CA), the membrane was processed for chemiluminescence with an Enhanced Chemiluminescence Western Blotting Detection Kit (Amersham Biosciences, Little Chalfont Buckinghamshire, England). Controls for loading were carried out after stripping the membrane (Restore, Invitrogen) using a mouse anti-actin antibody (Chemicon International, Temecula, CA) followed by an anti-mouse IgG HRP-conjugated secondary antibody (Novus Biologicals). Images were captured using a Lumiimager (Roche, Indianapolis, IN). Quantitation of protein ratios normalized to actin were calculated by densitometry using Lumianalyst 3.0 software (Roche) and expressed as arbitrary units of luminescence (BLU).
Statistical analysis
Statistical analysis was performed by Mann-Whitney non-parametric test and Student’s t test.
RESULTS
AZT-DNA incorporation
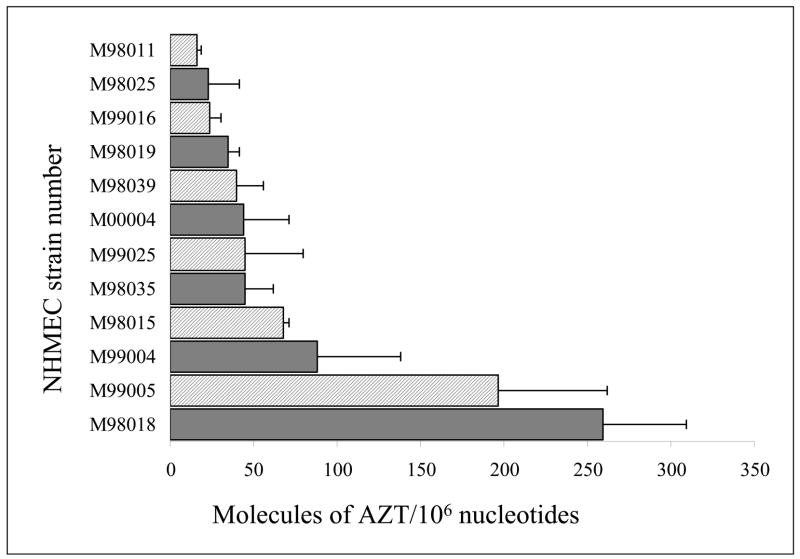
Twelve of 19 NHMEC strains exposed to 200 μM AZT for 24 h had detectable levels of AZT incorporation into DNA (Figure 1). Eight of the 19 NHMEC strains showed AZT-DNA incorporation levels ranging from 16 to 50 molecules of AZT/106 nucleotides. In two strains incorporation levels ranged from 51 to 100 molecules of AZT/106 nucleotides, while 2 strains had levels > 100 molecules of AZT/106 nucleotides. The remaining 7 NHMEC strains had no detectable AZT-DNA incorporation with an RIA limit of detection (LOD) of 16 molecules of AZT/106 nucleotides.
Figure 1.
Values for incorporation of AZT into DNA (molecules of AZT/106 nucleotides) are shown as bars for 12 NHMEC strains able to incorporate AZT into DNA after 24 h of exposure to 200 μM AZT. The 7 strains with undetectable levels of AZT-DNA incorporation at 24 h are: M98014, M98016, M98026, M98030, M98040, M99003 and M99015. DNA samples obtained from 2 separate cell exposures were each assayed 3 times by AZT-RIA, and values shown are mean ± range for n=2. The strains chosen for further analysis were: M99005, M98018, designated H1 and H2, respectively; and M98016 and M98040 designated L1 and L2, respectively.
Further analysis of genotoxicity was conducted in studies using two cell strains, H1 and H2 having high AZT-DNA incorporation at 24 h, and two strains L1 and L2, having undetectable levels of AZT-DNA incorporation at 24 h (Table 1). Cell viability at 24 h of exposure, assessed by cell counting, revealed similar survival values (81–85%) for strains H1, H2, L1 and L2 at 24 h of exposure to 200 μM AZT (Table 1). In order to examine AZT-DNA incorporation after longer incubations, strains L1 and L2 were exposed to AZT for an additional 48 h, and AZT-DNA incorporation was measured by RIA (Table 1). Cells from strain L1 had incorporation values of 95.0 ± 36.7 and 94.5 ± 15.7 molecules of AZT/106 nucleotides at 48 and 72 h of AZT exposure, respectively. Cells from strain L2 had 75.8 ± 19.1 molecules of AZT/106 nucleotides after AZT exposure for 48 h. Thus, it appears that some individuals showing no detectable AZT-DNA incorporation at 24 h have measurable AZT-DNA incorporation after incubation with AZT for 48 and 72 h.
Table 1.
Comparison of cell survival, AZT-DNA incorporation and % of cells in S-phase for NHMEC strains exposed to 200 μM AZT that incorporated the highest levels of AZT into DNA (H1 and H2), and those that incorporated low levels of AZT into DNA (L1 and L2).
| NHMEC Strain | Cell survival(%) at 24 ha | Molecules of AZT/106 nucleotidesb |
% of Cells in S-Phasec |
||
|---|---|---|---|---|---|
| 24 h | 48 h | No AZT | 200μM AZT | ||
| H1 | 84% | 197.0 ± 64.8 | NSd | 12.0 ± 4.4 | 18.0 ± 1.5 |
| H2 | 81% | 259.3 ± 18.5 | NS | 13.8 ± 0.2 | 19.4 ± 0.1f |
| L1 | 83% | NDe | 95.0 ± 36.7 | 9.3∀0.6 | 13.2 ± 1.1 |
| L2 | 85% | ND | 75.8 ± 19.1 | 17.3∀0.7 | 16.5 ± 0.8 |
Mean of 2 experiments each assayed twice by Trypan Blue.
Mean of 2 experiments each assayed three times by AZT-RIA.
Mean ± range of 2 experiments each assayed twice.
NS = no sample.
ND = not detectable; the lower limit of detection (LOD) was 16 molecules of AZT/106 nucleotides.
Difference between AZT-exposed and unexposed H2 cells was statistically significant (p<0.05).
BLM exposure and genotoxicity
In order to ascertain whether or not the variability of AZT-DNA incorporation values reflected a general response to DNA damage, or specific events occurring in response to AZT exposure, H1, H2, L1 and L2 cells were exposed to BLM, a direct-acting DNA damaging agent. BLM was chosen for these studies based on its capacity to inhibit incorporation of thymidine into DNA and to induce DNA double strand breaks, as both of these types of lesions are similar to DNA damage induced by AZT. Values for % of cells surviving at 24 h, after exposure to 36 μM BLM for 4 h, were 26%, 78%, 52% and 40% for strains H1, H2, L1 and L2, respectively, indicating that cell survival in BLM-exposed cells was not similar in the 4 strains. Furthermore, formation of micronuclei and apoptosis in BLM-exposed cells did not correlate with AZT-DNA incorporation (data not shown). Therefore, the lack of an intrinsic predisposition of NHMECs to process direct DNA damage in a manner that correlated with AZT-DNA incorporation, suggested that AZT-DNA interindividual variability is likely a specific response to AZT exposure.
Cell cycle alterations in AZT-exposed cells
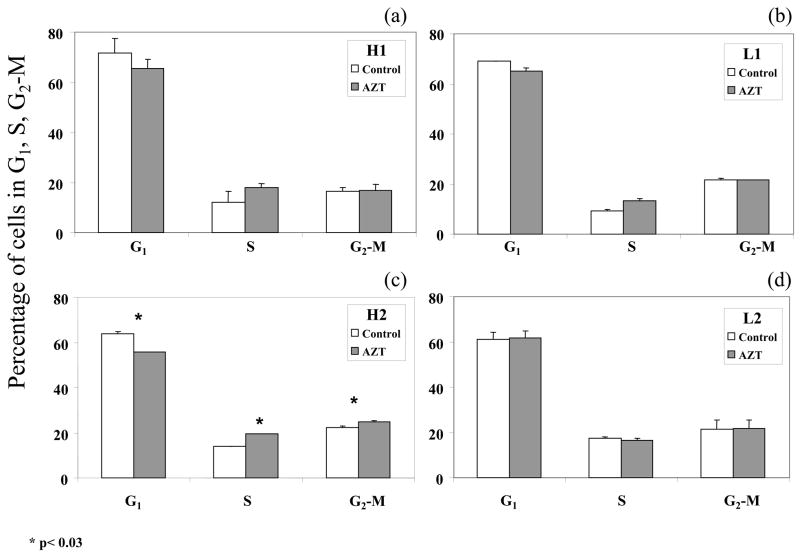
In order to further explore the genotoxic consequences of AZT-DNA incorporation in NHMEC strains H1, H2, L1 and L2, flow-cytometry was used to define cell cycle parameters after 24 h of exposure to 0 or 200 μM AZT. Two cell exposures were performed and, for each exposure, the cell cycle distribution was determined in duplicate. Separation of cells into G0-G1, S and G2-M phase of the cell cycle was based upon linear fluorescence intensity after staining with propidium iodide. Representative profiles (Figure 2) indicate the percentages of cells in G0-G1, S and G2-M phases of the cell cycle. AZT-induced changes in the cell cycle, suggesting accumulation of cells in S-phase, were observed in cell strains H1, H2 and L1 (Figure 2 a–c) but not in cell strain L2 (Figure 2d). AZT-induced increases in the percentages of cells in S-phase were 6.1%, 5.6% and 3.9% for strains H1, H2 and L1, respectively, but only strain H2 showed a statistically significant effect (Table 1 and Figure 2). Accompanying the AZT-induced accumulation of cells in S-phase was a decrease in the percentage of cells in G1-phase. Unexposed NHMEC H1, H2 and L1 cells had 71.6%, 63.9% and 69.1% of cells in G1, respectively, while exposed cells had 65.3%, 55.7% and 65.2% of cells in G1 (Figure 2, a, b and c). Student’s t test, was performed for all comparisons, and strain H2 showed statistically significant differences for comparison of unexposed and AZT-exposed cells in cell cycle phases G1, S and G2-M (Figure 2c). In contrast, strain L2 showed no change in % of cells in any cell cycle phase when AZT-exposed cells were compared with unexposed cells (Table 1 and Figure 2d).
Figure 2.
Percentages of cells in G1, S and G2-M phases of the cell cycle, determined by flow cytometry, in NHMECs H1, H2, L1 and L2 exposed to 0 (control, white bars) and 200μM AZT (grey bars) for 24 h. Each bar represents the mean plus range of duplicate readings from two independent experiments. Statistically-significant (p<0.03) differences between AZT-exposed and unexposed cells are indicated (*).
TK1 protein expression correlates with AZT-DNA incorporation
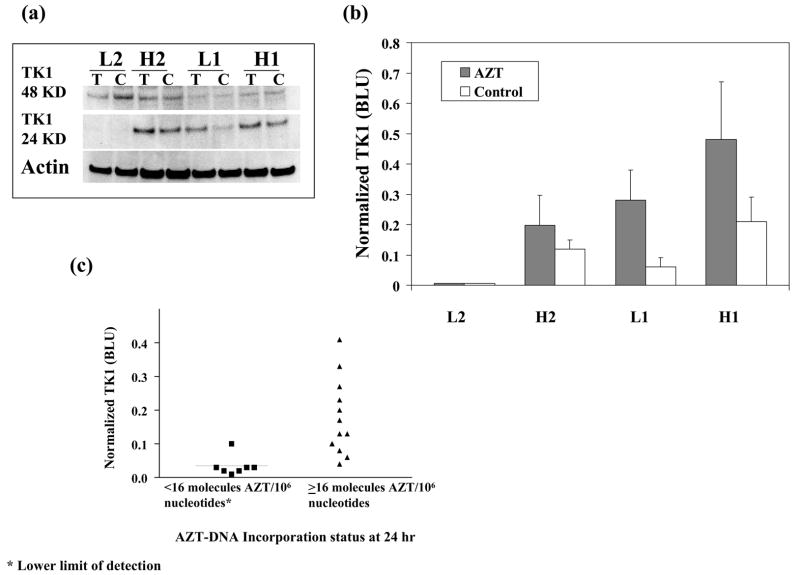
The wide range of AZT-DNA incorporation values shown for the 19 NHMEC strains (Figure 1) suggested that interindividual variability in AZT metabolism might contribute to these differences. Because the initial step in AZT metabolism, mono-phosphorylation, is accomplished by the monomeric (24 KD) active form of TK1, we examined TK1 protein levels using Western blot analysis. Figure 3a shows a representative protein blot for strains H1, H2, L1 and L2, with densitometric quantitation (normalized to actin) of the protein intensity for the same blot shown in Figure 3b. The induction of TK1 protein levels occurring as a result of AZT exposure in strains H1, H2 and L1 is shown in Figure 3, however strain L2, which had no measurable AZT-DNA at 24 h, also had a complete lack of constitutive or AZT-induced TK1 monomer at any time up to 24 h. Strain L2 also showed no AZT-DNA incorporation at 24 h and no AZT-induced changes in cell cycle parameters. The fact that TK1 is inducible in L1 but not L2 suggests more than one possible mechanism for the low levels of AZT-adducts measured in these two cell strains.
Figure 3.
(a) Representative Western blot, using a TK1 antibody, for cytoplasmic extracts of NHMECs H1, H2, L1 and L2 exposed to 0 (labeled C) or 200μM AZT (labeled T) for 24 h. The inactive dimeric (48 KD) and active monomeric (24 KD) forms of TK1 are shown in the upper rows, while the housekeeping protein actin is shown in the bottom row. (b) Normalized luminescence values for the data shown in Figure 3a, where the clear bars represent unexposed cells and the grey bars represent cells exposed to AZT. (c) Scatter plot for TK1 values (BLUs), obtained by normalized luminescence quantification, for the 19 NHMECs strains exposed to 200 μM AZT and grouped by capacity to incorporate AZT into DNA at 24 h. Cells with fewer than 16 molecules of AZT/106 nucleotides, the assay LOD, were grouped together and compared to those with ≥16 molecules of AZT/106 nucleotides. The mean BLU value (solid line on figure) ∀ SE for strains that have measurable AZT-DNA incorporation at 24 h was 0.18 ∀0.03 (n=12), compared to 0.03 ∀ 0.01 (n=7) for strains that have no measurable AZT-DNA incorporation at 24 h. The median BLU values were at 0.15 and 0.02 and the difference between the strains with ability to incorporate AZT (≥16 molecules of AZT/106 nucleotides) and those without it (<16 molecules of AZT/106 nucleotides) was statistically significant (p= 0.0005, Mann-Whitney test).
In a separate Western blot analysis the TK1 monomeric protein was quantified in all 19 NHMEC strains, for 24 h to 200 μM AZT. For the cells exposed to AZT (Figure 3c), the 7 NHMEC strains showing no AZT-DNA incorporation at 24 h had normalized TK1 protein values averaging 0.034 ± 0.01 BLUs (mean ± SE, n=7). In addition, the 12 NHMEC strains showing AZT-DNA incorporation at 24 h had normalized TK1 values of 0.18 ± 0.03 BLUs (mean ± SE, n=12) (Figure 3c). Overall, the active form of TK1 protein was significantly higher (p=0.0005) in AZT-exposed NHMECs able to incorporate AZT into DNA in 24 h, compared to those strains lacking that capacity. The data suggest that one major factor that determines variability in levels of AZT-DNA incorporation in NHMECs is the ability to up-regulate TK1 protein levels in response to AZT exposure.
DISCUSSION
This study has shown that widespread interindividual variability in incorporation of AZT into DNA is associated with the cellular capacity to induce TK1 in response to AZT exposure. Individuals showing the lowest, or no, incorporation of AZT into DNA at 24 h of exposure also showed no induction of TK1 protein in response to AZT exposure. TK1 performs the first phosphorylation of AZT and is considered a rate-limiting step in the pathway to nucleotide synthesis and DNA incorporation. The observation that TK1 protein levels influence the extent of AZT-DNA incorporation into human host cell suggests that patients with greater induction of TK1 and higher AZT-DNA incorporation levels may be at elevated risk for mutational events and health risks, and this possibility deserves further investigation.
A high degree of variability for incorporation of AZT into human leukocyte DNA was first reported by Olivero et al. (Olivero et al., 1999) who showed that in 28 HIV-1-infected adults, in 22 infants born to HIV-1-infected mothers, and in 11 HIV-1-infected mothers, paired with their HIV-1-uninfected infants, there was about a 20-fold variability in the range of values for AZT-DNA incorporation in either cord blood or peripheral blood. In the mother-infant pairs, there was no correlation for AZT-DNA level by pair or by duration of drug treatment during pregnancy (Olivero et al., 1999). Similarly, infant patas monkeys born to dams given human-equivalent NRTI dosing during gestation showed a large inter-individualindividual variability in AZT-DNA incorporation in many different organs taken at birth (Olivero et al., 2002). Given that AZT pharmacokinetics and metabolism are similar in all primates studied (Moodley et al., 1998; O’Sullivan et al., 1993; Patterson et al., 1997; Divi et al., 2007), the observed variability in AZT-DNA levels presented a curious finding. Perhaps the current study at least partially explains the previously-observed AZT-DNA variability as being caused by inter-individualindividual differences in TK1 inducibility in the presence of AZT.
In this study, we compared four NHMEC strains, two (H1 and H2) with the capacity to form high levels of AZT-DNA incorporation at 24 h, and two (L1 and L2) showing no AZT-DNA incorporation at 24 h. We found not only a correlation between the inducibility of TK1 in the presence of AZT and the capacity to incorporate the drug into DNA, but also an apparent weak but significant correlation between AZT-DNA incorporation and cell cycle arrest. In this study, the AZT-exposed H1 and H2 cells showed S-phase arrest evidenced by increases of 6.1% and 5.6%, respectively, for % of cells in S-phase. Both strains also showed decreases in % of cells in G1. However, only the H2 cells showed statistical significance (p <0.05). Compared to H1 and H2 cells, the AZT-exposed L1 cells showed a 3.9% increase in S-phase cells, and the L2 cells showed none at all. In previous studies, (Olivero et al., 2005) using cultured human tumor (HeLa) cells, higher doses of AZT were shown to induce an even stronger cell cycle arrest, with increases of 21.8% and 26.5% of cells in S-phase for doses of 250 and 500 μM AZT, respectively. Taken together, these studies suggest that the ability of a cell to induce cell cycle arrest in response to AZT exposure may be related to the capacity to damage DNA by AZT-DNA incorporation during DNA replication.
In this study, Western blot revealed a range of protein levels for the active TK1 monomer (24KD) induced in 19 different NHMEC strains in the presence of 200μM AZT, with the strains incorporating measurable AZT having the highest TK1 levels. The blots revealed the presence of an inactive 48 KD TK1 dimer, which did not correlate with the other end points measured. Whereas we do not know the basis for the observed differences in human AZT-related TK1 inducibility, there are a number of possibilities. For example, polymorphisms have been described (Murphy et al., 1986; Murphy et al., 1987). In addition, inhibition of TK1 activity by AZT exposure has been reported in T-lymphocytic cells (Avramis et al., 1993) and attributed to hypermethylation of the TK-1 promoter (Wu et al., 1995). Interestingly, variability in the response to AZT therapy has usually been considered due to virological, immunological or pharmacological factors (Fletcher, 1999), however, inhibition of TK1 enzyme has recently been reported to occur in HIV-1-infected patients (Turriziani et al., 2004; Jacobsson et al., 1995; Jacobsson et al., 1998) and it is possible that antiretroviral therapy may alter TK1 expression. Because the antiretroviral activity of AZT is so uniformly successful, it appears that even individuals who do not induce TK1 in response to AZT exposure have sufficient metabolic capacity for antiretroviral success.
In conclusion, here we have shown that the broad range of AZT-DNA incorporation values seen in 19 different NHMEC strains likely reflects human inter-individual variability in AZT-driven induction of TK1, the first enzyme along the pathway of AZT incorporation into DNA. Furthermore, the AZT-induced cell cycle arrest observed here in NHMECs and previously in HeLa cells appears to be associated with the ability of AZT to induce TK1 and become incorporated into DNA. It appears that the variations in TK1 metabolism observed here will not alter antiretroviral efficacy, however, the capacity to induce TK1 may be relevant to mechanisms of toxicity and the evolution of cellular resistance. For example, patients having high levels of TK1 and AZT-DNA incorporation may sustain more organ genotoxicity while those with low levels of TK1 and AZT-DNA incorporation may be less susceptible to host genotoxic effects.
Acknowledgments
All breast tissues, salvaged at reduction mammoplasty, were obtained through the cooperative Human Tissue Network (sponsored by the National Cancer Institute and the National Disease Research Interchange). This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
ABBREVIATIONS
- AZT
3′-azido-3′-deoxythymidine, Zidovudine
- BLM
Bleomycin
- BLU
Bright Luminescence Unit, an arbitrary unit of luminescence
- DAPI
4′,6-diamidino-2-phenylindole dihydrochloride
- ECL
Electrochemiluminescence
- FITC
Fluorescein isothiocyanate
- HRP
Horse Radish-peroxidase
- H1 and H2
NHMEC strains M99005 and M98018, respectively, the strains that incorporated the highest levels of AZT into DNA when measured by AZT-RIA, after 24 h of exposure to 200 μM AZT
- L1 and L2
NHMEC strains M98016 and M98040, respectively, two of the strains that incorporated undetectable levels of AZT into DNA when measured by AZT-RIA, after 24 h of exposure to 200 μM AZT
- NHMEC
Normal Human Mammary Epithelial Cell
- NRTI
Nucleoside reverse transcriptase inhibitor
- PBS
phosphate buffered saline
- RIA
Radioimmunoassay
- RIPA buffer
Radio-immune precipitation assay buffer
- TE
10 mM Tris, 1 mM EDTA buffer
- Tdt
Terminal deoxy-nucleotidyl transferase
- TK1
Thymidine kinase 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Avramis VI, Kwock R, Solorzano MM, Gomperts E. Evidence of in vitro development of drug resistance to azidothymidine in T-lymphocytic leukemia cell lines (Jurkat E6-1/AZT-100) and in pediatric patients with HIV-1 infection. JAcquir Immune Defic Syndr. 1993;6:1287–1296. [PubMed] [Google Scholar]
- Cardo DM, Culver DH, Ciesielski CA, Srivastava PU, Marcus R, Abiteboul D, Heptonstall J, Ippolito G, Lot F, McKibben PS, Bell DM. A case-control study of HIV seroconversion in health care workers after percutaneous exposure. Centers for Disease Control and Prevention Needlestick Surveillance Group. N Engl J Med. 1997;337:1485–1490. doi: 10.1056/NEJM199711203372101. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Department of Health and Human Services, Public Health Service, Centers for Disease Control and Prevention, National Institute for Occupational Safety and Health, DHHS (NIOSH); 1999. NIOSH alert: request for assistance in preventing needlestick injuries in health care settings. http://www.cdc.gov/niosh/2000-108.html. [Google Scholar]
- Centers for Disease Control and Prevention. Advancing HIV Prevention: the science behind the new initiative. 2003 http://www.cdc.gov/hiv/topics/prev.
- DHHS. Antiretroviral Guidelines for Adults and Adolescents - A Working Group of the Office of AIDS Research Advisory Council (OARAC) 2006 http://aidsinfo.nih.gov/contentfiles/AdultandAdolescentGL.pdf. DHHS Panel.
- Divi RL, Doerge DR, Twaddle NC, Shockley ME, St Claire MC, Harbaugh JW, Harbaugh SW, Poirier MC. Metabolism and pharmacokinetics of the combination Zidovudine plus Lamivudine in the adult Erythrocebus patas monkey determined by liquid chromatography-tandem mass spectrometric analysis. Toxicology and Applied Pharmacology. doi: 10.1016/j.taap.2007.09.007. http://dx.doi.org/10.1016/j.taap.2007.09.007. [DOI] [PubMed]
- Diwan BA, Riggs CW, Logsdon D, Haines DC, Olivero OA, Rice JM, Yuspa SH, Poirier MC, Anderson LM. Multiorgan transplacental and neonatal carcinogenicity of 3′-azido-3′-deoxythymidine in mice. Toxicol Appl Pharmacol. 1999;15:82–99. doi: 10.1006/taap.1999.8782. [DOI] [PubMed] [Google Scholar]
- Escobar PA, Olivero OA, Wade NA, Abrams EJ, Nesel CJ, Ness RB, Day RD, Day BW, Meng Q, O’Neill JP, Walker DM, Poirier MC, Walker VE, Bigbee WL. Genotoxicity assessed by the comet and GPA assays following in vitro exposure of human lymphoblastoid cells (H9) or perinatal exposure of mother-child pairs to AZT or AZT-3TC. Environ Mol Mutagen. 2007;48:330–343. doi: 10.1002/em.20285. [DOI] [PubMed] [Google Scholar]
- Fletcher CV. Pharmacologic considerations for therapeutic success with antiretroviral agents. Ann Pharmacother. 1999;33:989–995. doi: 10.1345/aph.19075. [DOI] [PubMed] [Google Scholar]
- Furman PA, Fyfe JA, St Clair MH, Weinhold K, Rideout JL, Freeman GA, Lehrman SN, Bolognesi DP, Broder S, Mitsuya H, Barry DA. Phosphorylation of 3′-azido-3′-deoxythymidine and selective interaction of the 5′-triphosphate with human immunodeficiency virus reverse transcriptase. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:8333–8337. doi: 10.1073/pnas.83.21.8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC. Some antiviral and antineoplastic drugs, and other pharmaceutical agents. Vol. 76. World Health Organization. International Agency for Research on Cancer; Lyon, France: 2000. Monographs on the Evaluation of Carcinogenic Risks to Humans; pp. 73–127. [Google Scholar]
- Jacobsson B, Britton S, He Q, Karlsson A, Eriksson S. Decreased thymidine kinase levels in peripheral blood cells from HIV-seropositive individuals: implications for zidovudine metabolism. AIDS Res Hum Retroviruses. 1995;11:805–811. doi: 10.1089/aid.1995.11.805. [DOI] [PubMed] [Google Scholar]
- Jacobsson B, Britton S, Tornevik Y, Eriksson S. Decrease in thymidylate kinase activity in peripheral blood mononuclear cells from HIV-infected individuals. Biochem Pharmacol. 1998;56:389–395. doi: 10.1016/s0006-2952(98)00032-x. [DOI] [PubMed] [Google Scholar]
- Keshava C, Whipkey D, Weston A. Transcriptional signatures of environmentally relevant exposures in normal human mammary epithelial cells: benzo[a]pyrene. Cancer Letters. 2005;221:201–211. doi: 10.1016/j.canlet.2004.08.037. [DOI] [PubMed] [Google Scholar]
- Moodley J, Moodley D, Pillay K, Coovadia H, Saba J, van Leeuwen R, Goodwin C, Harrigan PR, Moore KH, Stone C, Plumb R, Johnson MA. Pharmacokinetics and antiretroviral activity of lamivudine alone or when coadministered with zidovudine in human immunodeficiency virus type 1-infected pregnant women and their offspring. JInfect Dis. 1998;178:1327–1333. doi: 10.1086/314431. [DOI] [PubMed] [Google Scholar]
- Murphy PD, Kidd JR, Castiglione CM, Lin PF, Ruddle FH, Kidd KK. A frequent polymorphism for the cytosolic thymidine kinase gene, TK1, (17q21–q22) detected by the enzyme TaqI. Nucleic Acids Research. 1986;14:4381. doi: 10.1093/nar/14.10.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy PD, Lin PF, Ruddle FH, Kidd KK. A second useful polymorphism for the cytosolic thymidine kinase gene (TK1) with the enzyme BstEII which will allow haplotying at this locus on chromosome 17 (q21–q22) Nucleic Acids Research. 1987;15:7212. doi: 10.1093/nar/15.17.7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NTP. Toxicology and Carcinogenesis Studies of AZT. National Toxicology Program; Research Triangle Park, NC: 1996. [Google Scholar]
- O’Sullivan MJ, Boyer PJ, Scott GB, Parks WP, Weller S, Blum MR, Balsley J, Bryson YJ. The pharmacokinetics and safety of zidovudine in the third trimester of pregnancy for women infected with human immunodeficiency virus and their infants: phase I acquired immunodeficiency syndrome clinical trials group study (protocol 082). Zidovudine Collaborative Working Group. Am J Obstet Gynecol. 1993;168:1510–1516. doi: 10.1016/s0002-9378(11)90791-1. [DOI] [PubMed] [Google Scholar]
- Olivero OA. Mechanisms of genotoxicity of nucleoside reverse transcriptase inhibitors. Environ Mol Mutagen. 2007;48:215–223. doi: 10.1002/em.20195. [DOI] [PubMed] [Google Scholar]
- Olivero OA, Anderson LM, Diwan BA, Haines DC, Harbaugh SW, Moskal TJ, Jones AB, Rice JM, Riggs CW, Logsdon D, Yuspa SH, Poirier MC. Transplacental effects of 3′-azido-2′,3′-dideoxythymidine (AZT): tumorigenicity in mice and genotoxicity in mice and monkeys. J Natl Cancer Inst. 1997;89:1602–1608. doi: 10.1093/jnci/89.21.1602. [DOI] [PubMed] [Google Scholar]
- Olivero OA, Beland FA, Poirier MC. Immunofluorescent localization and quantitation of 3′-azido-2′, 3′-dideoxythymidine (AZT) incorporated into chromosomal DNA of human, hamster and mouse cell lines. International Journal of Oncology. 1994;4:49–54. doi: 10.3892/ijo.4.1.49. [DOI] [PubMed] [Google Scholar]
- Olivero OA, Fernandez JJ, Antiochos BB, Wagner JL, St Claire ME, Poirier MC. Transplacental genotoxicity of combined antiretroviral nucleoside analogue therapy in Erythrocebus patas monkeys. J Acquir Immune Defic Syndr. 2002;29:323–329. doi: 10.1097/00126334-200204010-00001. [DOI] [PubMed] [Google Scholar]
- Olivero OA, Shearer GM, Chougnet CA, Kovacs AA, Landay AL, Baker R, Stek AM, Khoury MM, Proia LA, Kessler HA, Sha BE, Tarone RE, Poirier MC. Incorporation of zidovudine into leukocyte DNA from HIV-1-positive adults and pregnant women, and cord blood from infants exposed in utero. AIDS. 1999;13:919–925. doi: 10.1097/00002030-199905280-00007. [DOI] [PubMed] [Google Scholar]
- Olivero OA, Tejera AM, Fernandez JJ, Taylor BJ, Das S, Divi RL, Poirier MC. Zidovudine induces S-phase arrest and cell cycle gene expression changes in human cells. Mutagenesis. 2005;20:139–146. doi: 10.1093/mutage/gei019. [DOI] [PubMed] [Google Scholar]
- Patterson TA, Binienda ZK, Lipe GW, Gillam MP, Slikker W, Jr, Sandberg JA. Transplacental pharmacokinetics and fetal distribution of azidothymidine, its glucuronide, and phosphorylated metabolites in late- term rhesus macaques after maternal infusion. Drug Metab Dispos. 1997;25:453–459. [PubMed] [Google Scholar]
- Poirier MC, Olivero OA, Walker DM, Walker VE. Perinatal genotoxicity and carcinogenicity of anti-retroviral nucleoside analog drugs. Toxicol Appl Pharmacol. 2004;199:151–161. doi: 10.1016/j.taap.2003.11.034. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor Laboratory Press; New York: 1989. [Google Scholar]
- Turriziani O, Antonelli G. Host factors and efficacy of antiretroviral treatment. New Microbiol. 2004;27:63–69. [PubMed] [Google Scholar]
- Vazquez IL, Olivero OA, Poirier MC. Altered AZT metabolism may induce cellular drug resistance in human cells. Environmental and Molecular Mutagenesis. 2004;44:234. Abstract # 186. [Google Scholar]
- Walker DM, Malarkey DE, Seilkop SK, Ruecker FA, Funk KA, Wolfe MJ, Treanor CP, Foley JF, Hahn FF, Hardisty JF, Walker VE. Transplacental carcinogenicity of 3′-azido-3′-deoxythymidine in B6C3F1 mice and F344 rats. Environ Mol Mutagen. 2007;48:283–298. doi: 10.1002/em.20297. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu X, Solorzano MM, Kwock R, Avramis VI. Development of zidovudine (AZT) resistance in Jurkat T cells is associated with decreased expression of the thymidine kinase (TK) gene and hypermethylation of the 5′ end of human TK gene. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;8:1–9. [PubMed] [Google Scholar]