Abstract
Resistance to organophosphorus (OP) insecticides is associated with decreased carboxylesterase activity in several insect species. It has been proposed that the resistance may be the result of a mutation in a carboxylesterase that simultaneously reduces its carboxylesterase activity and confers an OP hydrolase activity (the “mutant ali-esterase hypothesis”). In the sheep blowfly, Lucilia cuprina, the association is due to a change in a specific esterase isozyme, E3, which, in resistant flies, has a null phenotype on gels stained using standard carboxylesterase substrates. Here we show that an OP-resistant allele of the gene that encodes E3 differs at five amino acid replacement sites from a previously described OP-susceptible allele. Knowledge of the structure of a related enzyme (acetylcholinesterase) suggests that one of these substitutions (Gly137 → Asp) lies within the active site of the enzyme. The occurrence of this substitution is completely correlated with resistance across 15 isogenic strains. In vitro expression of two natural and two synthetic chimeric alleles shows that the Asp137 substitution alone is responsible for both the loss of E3’s carboxylesterase activity and the acquisition of a novel OP hydrolase activity. Modeling of Asp137 in the homologous position in acetylcholinesterase suggests that Asp137 may act as a base to orientate a water molecule in the appropriate position for hydrolysis of the phosphorylated enzyme intermediate.
Carboxyl/cholinesterases are a multi-gene family of enzymes that hydrolyze a diverse range of carboxylesters. Enzymes of this family are inhibited essentially irreversibly by organophosphorus (OP) insecticides and nerve gases (1, 2). These compounds rapidly undergo the first step of hydrolysis leading to a phosphorylated enzyme, but the second and final hydrolytic step to regenerate free enzymes is generally very slow. In particular, OPs phosphorylate and thereby inhibit acetylcholinesterase (AChE). This in turn inhibits the normal function of AChE, which is to reset the cholinergic system by hydrolyzing the neurotransmitter, acetylcholine (3).
Resistance to OP insecticides has been associated with both modifications of the target molecule, AChE, and increased levels of metabolism by glutathione S-transferases, cytochrome P450s, and carboxylesterases (4). Mutations in AChE have been found in resistant strains that render the enzyme less sensitive to inhibition (5). However, specific point mutations in metabolic enzymes have not been documented previously. Mutations variously associated with either increased or decreased carboxylesterase activities have been implicated, but the only mechanism resolved at a molecular level involves increased activities due to gene amplification (6). The resulting overproduced esterases act primarily as high-affinity “sponges” that are preferentially phosphorylated by a molar equivalent of OP (7–9). No cases of OP resistance associated with decreases in carboxylesterase activity have yet been defined at the molecular level, although the biochemistry associated with such resistance has been described at varying levels of detail for the Indian meal moth, Plodia interpunctella (10), the blowflies, Chrysomya putoria (11) and Lucilia cuprina (12), and the housefly, Musca domestica (12–18).
The correlation between low carboxylesterase activity and OP resistance was initially described in M. domestica using the aliphatic ester substrate, methyl butyrate, to measure what was termed “ali-esterase” activity. This activity was found to be lower in some strains resistant to various OPs compared with susceptible strains (13, 14). To explain this correlation it was suggested that in OP-resistant strains the major ali-esterase had mutated to enhance its ability to hydrolyze OPs, and in so doing had compromised its ability to hydrolyze carboxylesters (the “mutant ali-esterase hypothesis”) (12–18). Recent site-directed mutagenesis on human butyrylcholinesterase (BuChE) provides evidence that such a change in substrate specificity is possible. A Gly117 → His substitution in the oxyanion hole, which stabilizes the oxyanion formed in the transition state, resulted in an enzyme with enhanced OP turnover (19).
Many aspects of OP resistance in L. cuprina parallel low ali-esterase-associated OP resistance in M. domestica. Therefore, it has been proposed that resistance in both species may be caused by similar mechanisms and mutations (12, 20). In L. cuprina, high-level resistance to certain OPs is conferred by the Rop-1 locus (21). Flies homozygous for Rop-1 have a nonstaining phenotype for the carboxylesterase isozyme, E3, when α- and β-naphthyl acetate (α- and β-NA) are used as substrates, suggesting that Rop-1 may encode E3 (20, 22). Furthermore, E3 and Rop-1 map to the same region of chromosome 4 (20, 22, 23), the E3 nonstaining phenotype and the resistant allele of Rop-1 cosegregate in genetic crosses (24), and EMS-induced Rop-1 mutants also display the E3 nonstaining phenotype (25). Other evidence strongly suggests that E3 is homologous to the ali-esterase of M. domestica. Both these enzymes and associated resistance phenotypes map to equivalent regions of homologous chromosomes (26), the enzymes comigrate on electrophoresis, and each is the major contributor to ali-esterase activity in OP-susceptible strains (12). The OP-susceptible forms of both enzymes are rapidly phosphorylated by the active (“oxon” or P=O) form of OPs at a rate equal to or higher than the corresponding phosphorylation of AChE, a property suggested to be a prerequisite for a mutation to confer OP resistance (18, 27, 28). As in some resistant strains of M. domestica, a novel OP hydrolase activity has been detected in diazinon-resistant strains of L. cuprina (12, 29).
We have described the cloning and identification of the E3 gene (LcαE7) from the OP-susceptible LS2 strain of L. cuprina (27, 30). Here we describe the sequence differences between susceptible and resistant alleles and demonstrate a causal link between an amino acid substitution and the biochemical changes associated with resistance. We then use the known tertiary structure of AChE to produce a model for how this substitution might achieve the observed transformation in substrate specificity.
MATERIALS AND METHODS
Fly Strains.
Fifteen strains of L. cuprina were made isogenic for their fourth chromosomes by backcrossing to a balancer stock (22). The resistance status of each strain was determined by topical application of diazinon dissolved in 1 μl of acetone or 0.5 μl of kerosene onto the scutellum of 3- to 5-day-old adult females (31, 32). Most of these strains have previously been described with respect to several carboxylesterase and OP hydrolase activities (12). Flies were reared as described previously (26).
Nucleic Acid Procedures.
Reverse Transcriptase–PCR (RT-PCR) was used to amplify a cDNA allele of the E3 gene (LcαE7) from the diazinon-resistant Llandillo 103 strain. Total cellular RNA was prepared from 4-day-old adults by the method of Chirgwin et al. (33). Poly(A)+ RNA was prepared from 500 μg of total RNA using affinity chromatography on oligo(dT) cellulose (34). Oligo(dT)-primed cDNA was made from 1 μg of poly(A)+ mRNA using reverse transcriptase (Superscript II) as per the manufacturer’s (GIBCO/BRL) instructions.
The entire LcαE7 coding region was amplified from 200 ng of cDNA using the Lc743/5′ and Lc743/3′ primers (27). Reactions contained 100 pmol of each primer, 40 μM of each dNTP and 10 mM Tris⋅HCl (pH 8.8), 10 mM KCl, 0.002% Tween 20 (vol/vol), and 2 mM MgCl2. Six units of Taq polymerase (Perkin–Elmer) were added after five min at 97°C followed by 40 cycles of 35 s at 97°C, 1 min at 60°C, and 2 min at 72°C. A final extension at 72°C for 8 min was included. The 1.7-kb product was gel purified and cloned into the EcoRV cleavage site of the pBSK− plasmid vector (Stratagene). Three clones derived from independent amplification reactions were sequenced on both strands using primers described previously (27). These primers were used in dye-primer and dye-terminator sequencing reactions following the manufacturer’s (Applied Biosystems) instructions. A consensus sequence for the Llandillo 103 allele was constructed from sequence of three clones; each was derived from independent amplification reactions.
LcαE7 (103 bp), nucleotides 380–483 (27), which includes the region encoding Gly137 of the oxyanion hole, was sequenced for the 15 strains described above on both strands. Genomic DNA was prepared from either eggs (35) or adult flies (36). One microgram of genomic DNA was used as template in amplification reactions. Also included was 100 pmol of 7F1 and 7R4 primers (27), 0.2 mM of each dNTP, 10 mM Tris⋅HCl (pH 8.3), 50 mM KCl, and 1.5 mM MgCl2. After 3 min at 97°C, 2.5 units of Taq polymerase were added. An initial extension at 72°C was held for 2 min followed by 34 cycles of 97°C for 35 s, 55°C for 1 min, and 72°C for 1 min, with a final extension of 72°C for 10 min. The resulting 1-kb product was purified through QIAquick spin columns, following the manufacturer’s (Qiagen, Chatsworth, CA) instructions. Five hundred nanograms of this template was used in dye-terminator sequencing reactions with the primers 7R2 (27) and 7F1a (5′-AGCTAAATCCCGAAACTAAAC-3′) as described above.
Enzyme Expression and Activity Analysis.
A cDNA clone of the consensus Llandillo 103 sequence (pLc7L103) was constructed to contain the same 5′ leader and Kozak (37) sequence as the reference-susceptible clone, pLc732 (truncated 5′ leader but otherwise identical to Lc743) expressed previously (27). To test whether possible effects on activity were the result of only one of the substitutional differences in the resistant allele, two further cDNA clones were constructed. These artificial alleles of LcαE7 were made by ligating the 5′ SacI fragment (SacI site at nucleotide position 800 of LcαE7) from pLc7L103 into the equivalent position in the susceptible pLc732 clone and vice versa. This created one clone with only the Asp137 substitution on the susceptible allele background (pLc7Asp137) and another with Gly137 on the resistant background (pLc7Gly137). Recombinant Bacpac6 (CLONTECH) viruses containing pLc7L103, pLc7Asp137, and pLc7Gly137 were then constructed and recombinant protein collected as described previously (27). Carboxylesterase activity was estimated spectrophotometrically for three artificial ester substrates: α-naphthyl acetate (α-NA), p-nitrophenyl acetate (p-NPA), and methylthiobutyrate (MtB) as described previously (27). OP hydrolase activity was measured radiometrically using [14C]CVP as the substrate (12). Activities were also determined for both nonrecombinant, Bacpac6-infected and uninfected Sf9 cells to provide a measure of background hydrolysis.
RESULTS AND DISCUSSION
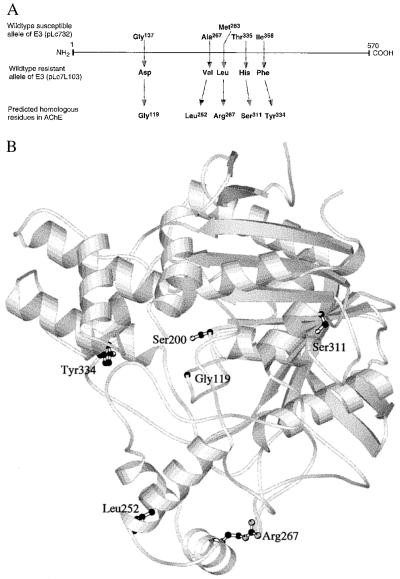
The nucleotide sequence of a diazinon-resistant Llandillo 103 allele of the LcαE7 gene encoding E3 differs from that of the OP-susceptible allele, LS2, described previously (27) at 19 nt sites, resulting in five amino acid substitutions: Gly137 → Asp, Ala267 → Val, Met283 → Leu, Thr335 → His, and Ile358 → Phe (Fig. 1A). Alignment of the E3 sequence with that of the related AChE from the electric ray, Torpedo californica (31% identical), for which the three-dimensional structure has been solved (39), predicts that only one of these five substitutions would be in the active site region of E3 (Fig. 1B). The residue homologous to Gly137 in AChE (Gly119) is 4.6 Å away from the γ oxygen of the nucleophile, Ser200, at the catalytic center of the molecule, whereas homologs of the other four substitutions (Leu252, Arg267, Ser311, Tyr334) are all at least 12 Å away. The γ oxygen of Ser200 is acylated in the first step of the hydrolytic reaction with carboxyl/cholinester substrates, or phosphorylated in the inhibition of the enzyme by OPs. Gly119 in AChE is part of the “oxyanion hole” that comprises three small residues (Gly118[136], Gly119[137], and Ala201[219] in AChE; E3 numbering in brackets) whose main chain nitrogens are thought to stabilize the oxyanions formed at the transition states in the two steps of the enzyme’s reaction mechanism (39). A glycine or other small amino acid is found at the position homologous to Gly119[137] in all catalytically active carboxyl/cholinesterases (1).
Figure 1.
(A) Amino acid differences between an OP-susceptible (pLc732) and a diazinon-resistant (pLc7L103) allele of E3. Predicted homologs in the amino acid sequence of Torpedo californica AChE are indicated below, based on a pileup alignment (38) modified to preserve secondary structure elements. (B) Three-dimensional structure of AChE (38) showing the side chains of the amino acids homologous to those that differ in the susceptible and resistant E3 alleles and the active site serine (Ser200) in ball-and-stick format. The rest of the molecule is depicted in ribbon form rendered using molscript (40).
Further evidence associating the Gly137 → Asp substitution in E3 with resistance was obtained by sequencing a 103-bp segment that includes the codon specifying Gly137 from 15 strains of L. cuprina drawn from widely distributed Australasian localities (Table 1). All seven diazinon-susceptible strains have a GGT triplet encoding Gly137, and all nine diazinon-resistant strains have a GAT encoding Asp137. No other amino acid replacement polymorphisms were found in this 103-bp segment of the E3 gene.
Table 1.
LD50 estimates for diazinon and identity of residue 137 of E3 for 15 isogenic (IV) strains of L. cuprina from across Australasia
| Strains | Origin of strain | LD50 diazinon, ng | Residue 137 of E3 |
|---|---|---|---|
| Diazinon susceptible | |||
| LS2 | Canberra, ACT, Australia | 40 | Gly |
| LBB 101 | Canberra, ACT, Australia | 50 | Gly |
| Llandillo 104 | Penrith, NSW, Australia | 50 | Gly |
| RM8 | Charleville, Qld, Australia | 80 | Gly |
| Woodside 5.2 | Adelaide, S. Aust., Australia | 50 | Gly |
| Hampton Hill 6.1 | Kalgoorlie, W. Aust., Australia | 20 | Gly |
| Hampton Hill 6.2 | Kalgoorlie, W. Aust., Australia | 30 | Gly |
| Diazinon resistant | |||
| Llandillo 103 | Penrith, NSW, Australia | 220 | Asp |
| RM2-6 | Charleville, Qld, Australia | 350 | Asp |
| Sunbury 5.2 | Boorowa, NSW, Australia | 220 | Asp |
| Gunning 107 | Gunning, NSW, Australia | 180 | Asp |
| Q4 | Northwest NSW, Australia | 290 | Asp |
| Strathfieldsaye 4.1 | Sale, Vic., Australia | 190 | Asp |
| Wanganui 5.3.3 | Wanganui, North Is., New Zealand | 240 | Asp |
| Wanganui 6.1.3 | Wanganui, North Is., New Zealand | 230 | Asp |
ACT, Australian Capital Territory; NSW, New South Wales; Qld, Queensland; S. Aust., South Australia; W. Aust., West Australia; Vic., Victoria; North Is., North Island.
To test whether only the Gly137 → Asp substitution in E3 is necessary to confer all the observed changes in biochemical phenotype of the enzyme, cDNA copies of four alleles of E3 were expressed in a baculovirus expression system (Fig. 2). Two were the naturally occurring wild-type susceptible (pLc732) and resistance (pLc7L103) alleles described above. A further two were synthetic chimeras, one with the Asp137 substitution transferred into the susceptible background (pLc7Asp137) and the other with the Gly137 substitution transferred into the resistant background (pLc7Gly137). The resistant enzyme and the chimera with the Asp137 on the susceptible background were able to hydrolyze the OP, CVP, but not the three artificial carboxylester substrates α-NA, p-NPA, and MtB. Conversely, the susceptible enzyme and the chimera with Gly137 on the resistant background could hydrolyze the carboxylesters but not CVP. Thus, the Asp137 substitution confers both biochemical changes associated with resistance that were linked by the mutant ali-esterase hypothesis, namely the loss of carboxylesterase activity and the acquisition of OP hydrolase activity. Given the parallels between OP resistance in L. cuprina and other insects (12), functionally equivalent mutations in E3 homologs are likely to explain other cases of resistance for which the mutant ali-esterase hypothesis has been invoked.
Figure 2.
Estimates of activity against three carboxylester (α-NA, p-NPA, MtB) and one OP (CVP) substrates for cell extracts containing baculovirus-expressed gene products of wild-type OP-susceptible LS2 strain (pLc732), wild-type diazinon-resistant Llandillo 103 strain (pLc7L103), and two artificially constructed chimeric alleles of E3 (pLc7Gly137, pLc7Asp137). Open circles represent residues found in the susceptible enzyme and solid circles represent those from the resistant enzyme. Nil indicates that there was no significant activity above those for uninfected cell extracts or extracts from nonrecombinant BacPac6-infected cells. All activity determinations are the mean of at least three estimates. In all cases activities two orders of magnitude less than the non-nil values shown would have been detected in these assays.
Although slow (turnover for diethyl OPs ≈0.5 min−1), this OP hydrolase activity is thought to be sufficient to protect AChE from inhibition by OPs and confer resistance (12, 16). The additional OP hydrolase activity observed in diazinon-resistant strains (≈23 pmol/min per fly; ref. 12) is sufficient to metabolize all of the additional dose that is tolerated by those strains (Table 1) in approximately 30 min. Devonshire and Moores (41) argued that turnover of 2.7 hr−1 for dimethyl OPs by the overexpressed esterase, E4, is sufficient to account for metabolism of a near-lethal dose in resistant strains of the aphid, Myzus persicae. Campbell et al. (12) argued that total metabolism of OPs in resistant strains of M. persicae and L. cuprina is similar. Whereas E4 represents up to 1% of the total soluble protein of M. persicae, E3 represents about 10-fold less of the soluble protein of a fly (28) but with about 10-fold higher turnover (41).
In some strains of housefly OP resistance is associated with elevated levels of cytochrome P450 and GST activities as well as decreased levels of carboxylesterase activity (42, 43). The factor responsible for conferring the elevated levels of these other metabolic enzymes maps close to the ali-esterase (44). Although there is no direct correlation between the mutant ali-esterase and elevated levels of GSTs or P450s in L. cuprina (45, 46), perhaps in M. domestica there is a second, linked resistance gene, or the mutant ali-esterase itself, that is responsible by some unknown mechanism for the up-regulation of these other metabolic enzymes in these resistant housefly strains (44).
OPs are hemisubstrates for carboxyl/cholinesterases. They form phosphorylated enzyme intermediates analogous to the acylated enzyme intermediates that form for carboxyl/cholinesters (2). However, the phosphorylated intermediate is impervious to the nucleophilic attack that normally completes the reaction for carboxyl/cholinester substrates. This is probably because of the different stereo-chemistries of the phosphoryl and carbonyl moieties (tetrahedral versus planar, respectively) that distinguish the two intermediates (47). Our finding of Asp137 in resistant E3 is consistent with the prediction that a mutation in this allele would place a nucleophilic residue in the appropriate position to activate a water molecule to attack the phosphorylated intermediate (12). We have modeled an aspartate residue in place of Gly119 on the known AChE structure and attached a diethylphosphate moiety to the active site serine (Ser200), with its phosphoryl oxygen oriented toward the oxyanion hole (Fig. 3). A water molecule, hydrogen bonded to the aspartate side chain, can be placed so that its oxygen atom could make a nucleophilic attack on the phosphorus atom in line with the serine–phosphorus bond. For carboxylester substrates the position of the Asp137 residue may compromise the ability of the oxyanion hole to stabilize the tetrahedral intermediate, explaining the loss of carboxylesterase activity in Asp137-E3.
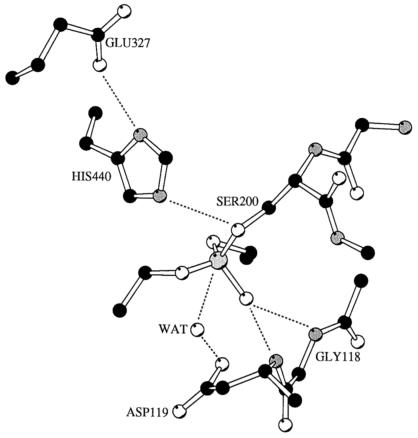
Figure 3.
Model of the active site region of AChE (39) with Gly119 (Gly137 in E3) substituted with an aspartate. Catalytic triad residues include Glu327, His440, and Ser200. Dashed lines indicate the positions of possible hydrogen bonds. A diethyl phosphate moiety has been attached to Ser200 to show the covalent intermediate that forms by the reaction of an OP with an esterase, and the position of a hypothetical water molecule (WAT) is indicated. The aspartate residue may act as a general base that enables water to attack the phosphorus atom, resulting in cleavage of the bond with Ser200. The direction of attack by the water molecule must be nearly in line with the bond to be cleaved (47, 48), and the model indicates that this is possible. The phosphoryl oxygen has been oriented toward the main chain nitrogen atoms of residues Gly118 and Asp119. Hydrogen bonds with these atoms may stabilize the oxyanion that would form during hydrolysis of the bond between Ser200 and the phosphorus atom. Modeling of the position of the water and Asp119 was conducted using van der Waals limitations and usual bond angles, and the model was rendered using molscript (40).
The substitution in the OP-hydrolyzing BuChE-His117 mutant (ref. 19; see Introduction) is in a homologous position to Gly137 of E3 and Gly119 in AChE. The mutant BuChE turns over diethyl OPs at the same rate as the mutant fly enzyme. For the mutant BuChE the authors propose two different mechanisms for OP hydrolysis in which His117 facilitates hydrolysis of the serine–phosphorus bond by a water molecule activated by His438 (His471 in E3, His440 in AChE) (19). Further biochemical and structural characterization will be required to differentiate among these various hypotheses. Whatever the mechanism of hydrolysis for either mutant, both are different from previously described OP hydrolases such as phosphotriesterases, which are metallo-enzymes and use a single-step reaction mechanism (48). For the insect enzyme, the evolution of OP resistance appears to have been achieved through a single mutational step, creating an OP hydrolase that is qualitatively different from both its parent enzyme and any other known phosphotriesterase.
Acknowledgments
We thank K. Medveczky for technical assistance and D. Beck, T. Boyce, C. Claudianos, R. Forster, D. Gleeson, D. Greenwood, B. Hammock, C. Robin, D. Rowell, M. Templeton, and two anonymous referees for discussions and comments on the manuscript. This work was supported by Australian Wool Research and Development (CEC 71), Horticulture Research and Development (HG 420), Rural Industries Research and Development (CSE 68A), and an Australian National University Ph.D. scholarship to R.D.N.
ABBREVIATIONS
- OP
organophosphate/organophosphorus
- AChE
acetylcholinesterase
- BuChE
butyrylcholinesterase
- α- and β-NA
α- and β-naphthyl acetate, respectively
- p-NPA
p-nitrophenyl acetate
- MtB
methylthiobutyrate
- CVP
chlorfenvinphos
References
- 1.Cygler M, Schrag J D, Sussman J C, Harel M, Silman M, Gentry K, Doctor B P. Protein Sci. 1993;2:366–382. doi: 10.1002/pro.5560020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldridge W N, Reiner E. Enzyme Inhibitors as Substrates: Interactions of Esterases with Esters of Organophosphorus and Carbamic Acids. Amsterdam: North–Holland; 1972. [Google Scholar]
- 3.Eto M. Organophosphorus Insecticides: Organic and Biological Chemistry. Cleveland: CRC; 1974. [Google Scholar]
- 4.Oppenoorth F J. In: Comprehensive Insect Physiology Biochemistry and Pharmacology. Kerkut G A, Gilbert L I, editors. Vol. 12. Oxford: Pergamon; 1985. pp. 731–773. [Google Scholar]
- 5.Mutero A, Pralavorio M, Bride J-M, Fournier D. Proc Natl Acad Sci USA. 1994;91:5922–5926. doi: 10.1073/pnas.91.13.5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devonshire A L, Field L M. Annu Rev Entomol. 1991;36:1–23. doi: 10.1146/annurev.en.36.010191.000245. [DOI] [PubMed] [Google Scholar]
- 7.Devonshire A L. Biochem J. 1977;167:675–683. doi: 10.1042/bj1670675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jayawardena K G I, Karunaratne S H P P, Ketterman A J, Hemingway J. Bull Ent Res. 1994;84:39–44. [Google Scholar]
- 9.Ketterman A J, Jayawardena K G I, Hemingway J. Biochem J. 1992;287:355–360. doi: 10.1042/bj2870355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beeman R W, Schmidt B A. J Econ Entomol. 1982;75:945–949. [Google Scholar]
- 11.Townsend M G, Busvine J R. Ent Exp Appl. 1969;12:243–267. [Google Scholar]
- 12.Campbell P M, Trott J F, Claudianos C, Smyth K-A, Russell R J, Oakeshott J G. Biochem Genet. 1997;53:17–40. doi: 10.1023/a:1022256412623. [DOI] [PubMed] [Google Scholar]
- 13.van Asperen K, Oppenoorth F J. Ent Exp Appl. 1959;2:48–57. [Google Scholar]
- 14.Bigley W, Plapp F. Ann Entomol Soc Am. 1960;53:360–364. [Google Scholar]
- 15.Oppenoorth F J, van Asperen K. Science. 1960;132:298–299. doi: 10.1126/science.132.3422.298. [DOI] [PubMed] [Google Scholar]
- 16.Oppenoorth F J, Van Asperen K. Ent Exp Appl. 1961;4:311–333. [Google Scholar]
- 17.Oppenoorth F J. Ent Exp Appl. 1959;2:304–319. [Google Scholar]
- 18.van Asperen K, Oppenoorth F J. Ent Exp Appl. 1960;3:68–83. [Google Scholar]
- 19.Lockridge O, Blong R M, Masson P, Froment M-T, Millard C B, Broomfield C A. Biochemistry. 1997;36:786–795. doi: 10.1021/bi961412g. [DOI] [PubMed] [Google Scholar]
- 20.Hughes P B, Raftos D A. Bull Ent Res. 1985;75:535–544. [Google Scholar]
- 21.Arnold J T A, Whitten M J. Bull Ent Res. 1976;66:561–568. [Google Scholar]
- 22.Parker A G, Russell R J, Delves A C, Oakeshott J G. Pestic Biochem Physiol. 1991;41:305–318. [Google Scholar]
- 23.Smyth K-A, Russell R J, Oakeshott J G. Biochem Genet. 1994;32:437–453. doi: 10.1007/BF00566064. [DOI] [PubMed] [Google Scholar]
- 24.Russell R J, Dumancic M M, Foster G G, Weller G L, Healy M J, Oakeshott J G. In: Insecticide Resistance as a Model System for Studying Molecular Evolution. Barker J S F, Starmer W T, MacIntyre R J, editors. New York: Plenum; 1989. pp. 263–314. [Google Scholar]
- 25.McKenzie J A, Parker A P, Yen J L. Genetics. 1992;130:613–620. doi: 10.1093/genetics/130.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weller G L, Foster G G. Genome. 1993;36:495–506. doi: 10.1139/g93-068. [DOI] [PubMed] [Google Scholar]
- 27.Newcomb R D, Campbell P M, Russell R J, Oakeshott J G. Insect Biochem Mol Biol. 1996;27:15–25. doi: 10.1016/s0965-1748(96)00065-3. [DOI] [PubMed] [Google Scholar]
- 28.Parker A G, Campbell P M, Spackman M E, Russell R J, Oakeshott J G. Pestic Biochem Physiol. 1996;55:85–99. doi: 10.1006/pest.1996.0038. [DOI] [PubMed] [Google Scholar]
- 29.Hughes P B, Devonshire A L. Pestic Biochem Physiol. 1982;18:289–297. [Google Scholar]
- 30.Newcomb R D, East P D, Russell R J, Oakeshott J G. Insect Mol Biol. 1996;5:211–216. doi: 10.1111/j.1365-2583.1996.tb00056.x. [DOI] [PubMed] [Google Scholar]
- 31.Smyth K-A. Ph.D. thesis. Canberra: Australian National University; 1994. [Google Scholar]
- 32.Gleeson D M, Barry S C, Heath A C G. Vet Parasitol. 1994;53:301–308. doi: 10.1016/0304-4017(94)90194-5. [DOI] [PubMed] [Google Scholar]
- 33.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 35.Davis L G, Dibner M D, Battey J F. Basic Methods in Molecular Biology. New York: Elsevier; 1986. [Google Scholar]
- 36.Crozier Y C, Koulianos S, Crozier R H. Experientia. 1991;47:968–969. doi: 10.1007/BF01929894. [DOI] [PubMed] [Google Scholar]
- 37.Kozak M. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sussman J S, Harel M, Frolov F, Oefner C, Goldman A, Toker L, Silman I. Science. 1991;253:872–879. doi: 10.1126/science.1678899. [DOI] [PubMed] [Google Scholar]
- 40.Kraulis P J. J Appl Cryst. 1991;24:946–950. [Google Scholar]
- 41.Devonshire A L, Moores G D. In: Enzymes Hydrolysing Organophosphorus Compounds. Reiner E, Aldridge W N, Hoskin F C G, editors. Chichester, U.K.: Ellis Horwood; 1989. pp. 180–192. [Google Scholar]
- 42.Carino F, Koener J F, Plapp F W, Feyereisen R. Insect Biochem Mol Biol. 1994;24:411–418. doi: 10.1016/0965-1748(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 43.Syvanen M, Zhou Z, Wang J. Mol Gen Genet. 1994;245:25–31. doi: 10.1007/BF00279747. [DOI] [PubMed] [Google Scholar]
- 44.Plapp F W. Pestic Biochem Physiol. 1984;22:194–201. [Google Scholar]
- 45.Board P, Russell R J, Marano R J, Oakeshott J G. Biochem J. 1994;299:425–430. doi: 10.1042/bj2990425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson J A, Clarke A G. Pestic Biochem Physiol. 1996;54:85–95. [Google Scholar]
- 47.Järv J. In: Enzymes Hydrolysing Organophosphorus Compounds. Reiner E, Aldridge W N, Hoskin F C G, editors. Chichester, U.K.: Ellis Horwood; 1989. pp. 221–225. [Google Scholar]
- 48.Lewis V E, Donarski W J, Wild J R, Raushel F M. Biochemistry. 1988;27:1591–1597. doi: 10.1021/bi00405a030. [DOI] [PubMed] [Google Scholar]